Mice
C57BL/6 (WT, CD45.2) and C57BL/6 CD45.1 mice were purchased from Jackson Laboratory. Il1rl1−/− (ST2 deficient) and Il33−/− mice were a gift from M. J. Rosen. Ltbfl/fl mice and Ltbr−/− mice were previously described51,52. Il7rcre/+Rorafl/fl mice were a gift from A. N. J. McKenzie. Nmur1icre-eGFPRosaLSL-DTR mice were previously described20. Ltbfl/fl mice were crossed to Nmur1icre-eGFP and Il7rcre/+ mice to obtain Nmur1icre-iGFPLtbfl/fl and Il7rcre/+Ltbfl/fl mice. CAG-KikGR53 mice were a gift from G. E. Diehl. Germ-free mice were provided by the gnotobiotic facility at Weill Cornell Medical Center. For all experiments, 6–14-week-old mice were age and sex matched and randomly assigned to specified treatment groups, with at least two independent experiments performed throughout. Sample sizes for experiments were determined without formal power calculations. Animals were bred and maintained in a specific-pathogen-free animal facility. All experiments were conducted in accordance with an Institutional Animal Care and Use Committee (IACUC) approved protocol at Memorial Sloan Kettering Cancer Center (MSK) and in compliance with all IACUC-relevant ethical regulations.
Cell lines and animal procedures
Cell lines
All tumour cell lines (gifts from R. H. Vonderheide and B. J Stanger) were derived from KPC (Pdx1-cre;LSL-KrasG12D/+;LSL-Trp53R172H/+) or KPCY (Pdx1-cre;LSL-KrasG12D/+;LSL-Trp53R172H/+;Rosa26YFP/YFP) mice. All cell lines were authenticated as bona fide PDAC cell lines based on histopathological verification by a dedicated pancreatic cancer pathologist. The HEK-Blue-IL-33 cell line (Invivogen) was cultured in DMEM (Gibco), 10% FBS (Gibco), penicillin (100 IU ml−1), streptomycin (100 µg ml−1) and 100 µg ml−1 normocin (Invivogen) at 37 °C in 5% CO2. All of the other cell lines were cultured in DMEM with 10% FBS and glutamine (2 mM) at 37 °C in 5% CO2. All of the cell lines were regularly tested using the MycoAlert Mycoplasma Detection Kit (Lonza).
PDAC tumours
Tumours were implanted orthotopically (pancreatic, PDAC mice) or s.c. as previously described16. In brief, for orthotopic implantation, mice were anaesthetized using isoflurane, and a small (7 mm) left abdominal side incision was made. Tumour cells (106 cells for KPC-4662 (PDAC 6); 105 cells for all others) were suspended in Matrigel (Becton Dickinson), diluted 1:1 with cold PBS, and injected as a 50 µl volume into the tail of the pancreas with a 31-gauge needle. Successful injection was verified by the appearance of a fluid bubble without intraperitoneal (i.p.) leakage. The abdominal wall was closed with absorbable PDS-II sutures (Ethicon), and the skin was closed with wound clips (Roboz). For s.c. implantation, tumour cells (5 × 105 cells for KPC-4662 (PDAC 6); 5 × 104 cells for all others) were resuspended in sterile PBS and implanted s.c. All tumours (PDAC and s.c.) were established with KPC-4662 (PDAC 6) unless otherwise specified. For sham surgery, mice were treated with orthotopic Matrigel injections only.
In PDAC mice, tumour volumes were measured using serial ultrasound (Vevo 2100 microultrasound, Visual Sonics) and analysed (Vivo Lab v.3.1.1, Fuji Film Visual Sonics) as previously described16. Tumours were collected at the indicated timepoints. To assess T cell infiltrates in PDACs derived from different cell lines (Extended Data Fig. 2c), tumours were collected at timepoints with comparable volumes. For s.c. tumours, tumour length and width were measured every 2–3 days with callipers, and tumour volumes were calculated as volume = (length/2) × width2. Mice were euthanized at the indicated timepoints and processed for histology or flow cytometry. No blinding was performed in experimental mouse interventions, as knowledge of the treatment groups was required. For survival analyses, survival was determined by a tumour volume of ≥500 mm3 or mouse health requiring euthanasia as defined by institutional IACUC guidelines. No mouse tumours exceeded IACUC-defined maximal tumour volumes of ≥2 cm3. In Fig. 2b and Extended Data Figs. 2c,d and 3e, PDAC 1 is cell line 6419, PDAC 2 is cell line 50, PDAC 3 is cell line 6694, PDAC 4 is cell line 52, PDAC 5 is cell line 6499 and PDAC 6 is cell line 4662.
Anti-LTβR treatment
Mice were treated with 100 µg of anti-LTβR agonistic antibody (3C8, Invitrogen) or isotype control (Invitrogen) i.p. every 3 days as described54.
DSS-colitis
3% DSS (m/v) (MP Biomedicals) was dissolved in drinking water. Mice were iteratively exposed to DSS for 7 days, followed by 7 days of recovery55, and euthanized for analyses at the indicated experimental timepoints. rIL-33 was initiated on day 0 of DSS treatment.
Acute ILC2 depletion with diphtheria toxin
Nmur1icre-eGFPRosaLSL-DTR mice or littermate control RosaLSL-DTR mice were treated with twice daily i.p. injections of 100 ng DT (EMD Millipore) for 7 days beginning 2 days before tumour implantation, and every 2 days thereafter, as previously described56.
Parabiosis
Female congenic CD45.1 and CD45.2 mice (aged 6 weeks) were surgically connected as previously described32. In brief, mice of similar body weight were co-housed 2 weeks before surgery and placed onto continuous prophylactic antibiotics (Sulfatrim diet, WF Fisher and Son) beginning the day before surgery. Lateral skin incisions from the elbow to knee were made on each mouse, and forelimbs and hindlimbs were sutured together. After surgery, mice were maintained on prophylactic sulfamethoxazole (Sulfatrim diet) for 2 weeks, followed by a normal diet thereafter. After confirming blood chimerism 4 weeks following parabiotic surgery, pancreatic and/or s.c. PDACs were implanted as described above. To ablate microbiota in parabiotic mice, after confirming blood chimerism, donor mice were treated with daily antibiotics (as described below) by oral gavage for 5 days before PDAC implantation, with subsequent antibiotic treatment maintained in the drinking water throughout the experimental duration. Parabionts were euthanized and organs were collected 14 days after tumour implantation.
Photoconversion
ILC2 migration was tracked using CAG-KikGR57 mice that express Kikume Green-Red (KikGR) photoconvertible fluorescent protein. In brief, orthotopic PDACs were established, PDAC mice were treated daily with rIL-33 and, 6 days after tumour implantation, tissue was photoconverted53. To do so, a 1 cm incision was made into the abdominal wall, and the caecum and small intestine (gut) or peritoneum were photoconverted using a 405 nm violet laser for 10 min. PDACs were collected 2 days later to assess gut-derived photoconverted ILC2s by flow cytometry.
Microbiome ablation
Microbiome was ablated using an antibiotic cocktail of vancomycin (50 mg ml−1; Sigma-Aldrich), neomycin (10 mg ml−1; Sigma-Aldrich), metronidazole (100 mg ml−1; Santa Cruz Biotech) and amphotericin (1 mg ml−1; MP Biomedicals) administered daily by oral gavage for 5 days36. Orthotopic PDAC tumours were then implanted, and microbiome ablation was maintained with ampicillin (1 mg ml−1; Sigma-Aldrich), vancomycin (0.5 mg ml−1; Sigma-Aldrich), neomycin (0.5 mg ml−1; Sigma-Aldrich), metronidazole (1 mg ml−1; Santa Cruz Biotech) and amphotericin (0.5 μg ml−1; MP Biomedicals) in the drinking water for the duration of the experiment.
Faecal microbiota transfer
To reconstitute microbiota in germ-free mice, fresh faecal samples were collected from WT C57BL/6 mice, resuspended in sterile PBS (6 faecal pellets per 1 ml of PBS), and 200 µl of the suspension was administered by oral gavage every other day for 2 weeks36. PDACs were then implanted and microbiome reconstitution was maintained with subsequent weekly faecal microbiota transfer.
Blood collection
Peripheral blood was collected from the submandibular vein of mice using a golden rod animal lancet (Medipoint) in plasma separation tubes with lithium heparin (BD Biosciences) and Microvette CB300Z (Sarstedt).
Microbiome analysis
DNA extraction and 16S library generation
Faecal samples were collected, DNA was extracted from single faecal pellets and deposited into a Qiagen PowerBead glass 0.1 mm tube (Qiagen) using the Promega Maxwell RSC PureFood GMO and Authentication Kit (Promega) according to the manufacturer’s instructions. DNA was quantified using the Quant-iT dsDNA High Sensitivity Assay Kit with the Promega GloMax plate reader on a microplate (Fisher Scientific). 16S libraries were then generated as described previously (Earth Microbiome Project; https://earthmicrobiome.org/protocols-and-standards/16s/). Amplicon libraries were washed using Beckman Coulter AMPure XP magnetic beads, and the library quality and size were verified using the PerkinElmer LabChip GXII with the DNA 1K Reagent Kit (Perkin Elmer) according to the manufacturer’s instructions. Libraries were normalized to 2 nM using the PerkinElmer Zephyr G3 NGS Workstation (Perkin Elmer) and pooled using identical volumes across normalized libraries into a 1.5 ml Eppendorf DNA tube.
Sequencing and data processing
Pooled libraries were sequenced (Illumina MiSeq) at a loading concentration of 7 pM with 15% PhiX, paired-end 250 (MiSeq Reagent Kit v2, 500-cycles). Demultiplexed raw reads were processed using the Nextflow58, nf-core59 ampliseq60 pipeline (v.2.4.0), with the following parameters: -profile singularity –FW_primer GTGYCAGCMGCCGCGGTAA –RV_primer CCGYCAATTYMTTTRAGTTT –dada_ref_taxonomy silva –ignore_empty_input_files –ignore_failed_trimming –min_frequency 10 –retain_untrimmed –trunclenf 240 –trunclenr 160. Specifically, reads were trimmed with cutadapt61, PhiX, and quality filtering, read pair merging and amplicon sequence variant resolution was performed with DADA262. Subsequent taxonomic assignment was also performed with DADA2, using the Silva reference database63 (v.138). Abundance tables were analysed using the QIIME2 software package. Tables were rarefied to 8,000 sequences for the Shannon diversity and principal component analyses.
Recombinant H-rIL-33 and H-e-rIL-33–Fc
H-rIL-33 and H-e-rIL-33–Fc proteins were produced by GenScript Biotech. In brief, target DNA sequences were codon-optimized, synthesized and subcloned into a cytomegalovirus promoter-driven expression vector following the human IL-2 signal peptide sequence. The proteins were expressed by transient transfection in HD Chinese hamster ovary cells and purified by affinity chromatography, followed by size-exclusion chromatography to obtain the desired purity. The purified protein was analysed by SDS–PAGE, western blotting and high-performance liquid chromatography analysis to determine the molecular mass and purity.
Recombinant IL-33, IL-25, H-rIL-33 and H-e-rIL-33–Fc treatment
After tumour implantation, mice were treated with i.p. injections of 500 ng carrier-free recombinant murine IL-3316, IL-25 (R&D Systems) or H-rIL-33 (Proteos) daily for 7 days, and then every 2 days thereafter. For H-e-rIL-33–Fc, after tumour implantation, mice were treated every 2 days for 7 days, and then every 4 days thereafter at the indicated doses.
Human samples
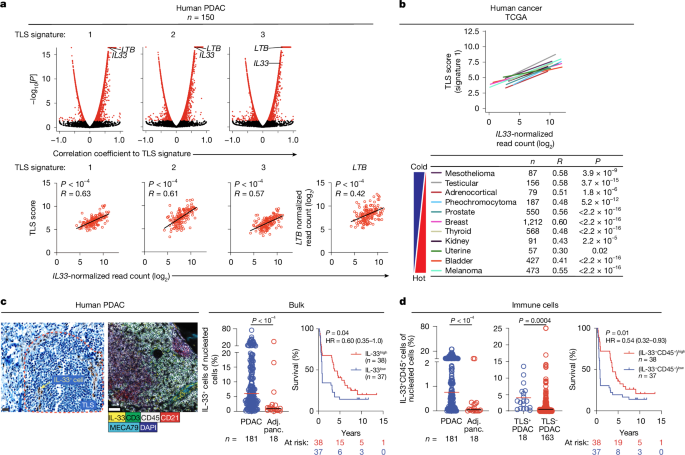
All tissues were collected at MSK according to MSK Institutional Review Board (IRB) approved study protocols. Informed consent was obtained for all of the patients. The study was performed in strict compliance with all institutional ethical regulations. All tumour samples were surgically resected primary PDACs (for tumour transcriptomic profiling and serum IL-33 measurement; Supplementary Tables 1 and 3), or surgically resected human PDAC (immunohistochemistry/immunofluorescence, flow cytometry; Supplementary Tables 2 and 5). The human PDAC tissue microarrays16, PDAC RNA-seq from ICGC64 and human tumour sequencing from TCGA65 have been previously described.
Tumour transcriptomic profiling
For the MSK cohort, primary PDACs from surgically resected patients with PDAC were randomly selected to undergo transcriptomic profiling as previously described16. In brief, total RNA from fresh-frozen OCT-embedded tumours was extracted using TRIzol RNA Isolation Reagents (Life Technologies), qualified on an Agilent BioAnalyzer, quantified by fluorometry (Ribogreen) and prepared for whole-transcriptome expression analysis using the WT Pico Reagent Kit (Affymetrix). RNA was then amplified using low-cycle PCR followed by linear amplification using T7 in vitro transcription technology. The cRNA was then converted to biotinylated sense-strand DNA hybridization targets, hybridized to GeneChip Human Transcriptome Array 2.0 (Affymetrix), scanned using the GeneChip Scanner 3000 and analysed using R (v.4.0.3).
For the TCGA cohort, RNA-seq datasets were obtained from https://gdc.cancer.gov/ under the identifiers of TCGA-PAAD66 (TCGA PDAC cohort), TCGA-BRCA67, TCGA-SKCM68, TCGA-ACC, TCGA-BLCA, TCGA-KICH, TCGA-MESO, TCGA-PCPG, TCGA-PRAD, TCGA-TGCT or TCGA-UCS. For the ICGC cohort, RNA-seq data were obtained from previously published data64. For PDACs in all cohorts, patients with histologically diagnosed PDAC were included. All data were log2-transformed.
Cell isolation
Mouse and human PDACs and mouse intestines were mechanically dissociated and incubated in collagenase (collagenase II for mouse tumours, collagenase IV for human tumours, both 5 mg ml−1; Worthington Biochemical, Thermo Fisher Scientific), DNase I (0.5 mg ml−1; Roche Diagnostics) and Hank’s balanced salt solution (Gibco, Thermo Fisher Scientific) for 30 min at 37 °C. Digestion was then quenched with FBS (Life Technologies). Digested tumours and DLNs were then mechanically disassociated and filtered through 100 mm and 40 mm nylon cell strainers (Falcon, Thermo Fisher Scientific) using PBS with 5% FBS (Life Technologies) and 4 mM EDTA (pH 8.0, Invitrogen). Spleens were mechanically dissociated and filtered through 70 mm and 40 mm nylon cell strainers (Falcon, Thermo Fisher Scientific) using PBS with 5% FBS and 2 mM EDTA, followed by red blood cell (RBC) lysis (RBC lysis buffer, Invitrogen Scientific). Peripheral blood was processed with RBC lysis and filtered through 40 mm nylon cell strainers. Mouse Fc receptors were blocked with FcεRIII/II-specific antibody (1 µg per 1 × 106 cells; 2.4G2, Bio XCell).
ILC2 and myeloid cell adoptive transfer
Donor orthotopic PDAC donor mice were treated with 500 ng carrier-free murine rIL-33 (R&D Systems) in sterile PBS daily for 10 days. For ILC2 transfer, live CD45+lin−CD90+KLRG1+ ILC2s from tumours were sort-purified to 98% purity at day 10 after implantation using the Aria Cell Sorter (BD Biosciences). Then, 5 × 104 ILC2s were immediately transferred to orthotopic PDAC-tumour-bearing Il7rcre/+Rorafl/fl mice through i.p. injection 3 days after tumour implantation. For myeloid cell transfer, live CD45+NK1.1−CD11b+LTβR+ cells were sort-purified to 90% purity using the Aria Cell Sorter. For adoptive transfer, 5 × 105 CD11b+LTβR+ cells were mixed with tumour cell suspensions and injected into the tail of the pancreas using a 31-gauge needle. rIL-33 treatment (500 ng per mouse as described above) was administered in recipient mice on the day of ILC2 or myeloid cell transfer until the day of tissue collection. Tissues were collected at the indicated timepoints.
Flow cytometry
All of the samples were analysed on the FACS LSR Fortessa (BD Biosciences) system. Mouse ILC2s were defined as live CD45+lin−(CD3, CD5, NK1.1, CD11b, CD11c, CD19, FcεR1)CD90+. Mouse KLRG1+ ILC2s were defined as live CD45+lin−(CD3, CD5, NK1.1, CD11b, CD11c, CD19, FcεR1)CD90+KLRG1+. All KLRG1+ ILC2s as defined were confirmed to be GATA3+ in our model (Extended Data Fig. 3a). Human ILC2s were defined as live CD45+lin−(CD3, CD5, CD56, CD11b, CD11c, CD14, CD16, CD19, TCRα/β, FcεR1)CD127+CRTH2+. Human KLRG1+ ILC2s were defined as live CD45+lin−(CD3, CD5, CD56, CD11b, CD11c, CD14, CD16, CD19, TCRα/β, FcεR1)CD127+CRTH2+KLRG1+. The following definitions were used for other immune cells: ILC1, live CD45+lin−TBET+; ILC3, live CD45+lin−NK1.1−RORγt+; NK, live CD45+CD3−NK1.1+; B cells, live CD45+CD3−NK1.1−CD19+; CD4+ T cells, live CD45+NK1.1−CD3+CD8−CD4+; CD8+ T cells, live CD45+NK1.1−CD3+CD4−CD8+; Eosinophils, live CD45+NK1.1−CD3−CD19−CD11b+SIGLECF+; macrophages, live CD45+NK1.1−CD3−CD19−CD11b+F4/80+; myeloid-derived suppressor cells (MDSCs), live CD45+NK1.1−CD3−CD19−CD11b+SiglecF−Gr-1+; DCs, live CD45+NK1.1−CD3−CD19−Gr-1−F4/80−CD11c+MHC-II+.
Mouse cells were stained with the following antibodies according to the manufacturer’s instructions: from BioLegend, CD11c (N418, BV510), CD19 (6D5, PE and BV785), CD25 (PC61, PerCP-Cy5.5), CD3 (145-2C11, BV711), CD4 (RM4-5, BV711 and BV786), CD45 (30-F11, Pacific Blue), CD45.1 (A20, BV711 and APC-Cy7), CD45.2 (104, Pacific Blue and APC-Cy7), CD8 (53-6.7, BV510), KLRG1 (2F1/KLRG1, BV510), LTBR (5G11, PE-Cy7), NK1.1 (PK136, BV605), SIGLECF (S17007L, APC), CD127 (A7R34, PE-Cy7) and Zombie Red Fixable Viability dye (423110); from BD Biosciences, CD11b (M1/70, Alexa Fluor 700, APC, and APC-Cy7), CD5 (53-7.3, APC), CD11c (HL3, APC), CD90.2 (53-2.1, BV786), GATA3 (L50-823, BV711 and PE), Gr-1 (RB6-8C5, BV605), NK1.1 (PK136, BV650 and APC), DRAQ7 (51-9011172), T-BET (O4-46, BV650), RORγt (Q31378, PE) and 4′,6-diamidino-2-phenylindole (DAPI); from Invitrogen Scientific, CD11c (N418, FITC), CD127 (A7R34, FITC), CD19 (eBio1D3, Alexa Fluor 700), CD3 (17A2, Alexa Fluor 700), CD8 (53-6.7, Alexa Fluor 700), F4/80 (BM8, PE-Cy5), FceR1 (MAR-1, APC), FOXP3 (FJK-16s, FITC), IL-33 (396118, PE), MHC class II (M5/114.15.2, Alexa Fluor 700), ST2 (RMST2-2, PE-Cy7) and TCRVβ (MR-9-4, Alexa Fluor 700).
To detect mouse LT, single-cell suspensions were incubated with recombinant mouse LTβR–Fc chimeric protein (1 µg ml−1, R&D systems) in PBS with 5% FBS and 4 mM EDTA for 30 min in the dark at 4 °C, followed by incubation with a secondary antibody (goat anti-mouse IgG2a conjugated, Invitrogen) for 30 min in the dark at 4 °C, as previously described69. As positive controls, 3 × 106 bulk splenic T cells (isolated by negative selection using the mouse Pan T Cell Isolation Kit II, Miltenyi Biotech) from WT (positive control) or Ltb−/− (negative control) mice were stimulated in vitro with anti-CD3/anti-CD28 (1 μg ml−1, BD Biosciences) in coated six-well plates with 10% RPM, 100 IU ml−1 penicillin and 100 µg ml−1 streptomycin for 4 h based on a previously described protocol70 and examined for surface LT expression using identical staining methods as described above. To analyse mouse NMUR1 protein expression, single-cell suspension was first blocked with 2% normal mouse serum (Jackson ImmunoResearch) in FACS buffer (1% BSA, 2.5 mM EDTA, 25 mM HEPES, 0.05% sodium azide in PBS) followed by viability staining using LIVE/DEAD Fixable Aqua Dead Cell Stain dye (Thermo Fisher Scientific) in PBS. Cells were then incubated 30 min with anti-mouse NMUR1-biotinylated antibody (12-A03-A; previously described)20 at 0.0173 mg ml−1 contained in the surface staining antibody mix. Then, after washing with FACS buffer, cells were counterstained with Streptavidin-BV650 conjugated fluorochrome at 1/250 dilution (BioLegend) for 15 min, followed by the intracellular staining and detection using the methods described above.
Human cells were stained with the following antibodies according to the manufacturer’s instructions: from BD Biosciences, GATA3 (L50-823, PE); from BioLegend, CD11b (ICRF44, APC), CD45 (HI30, Pacific Blue), CD56 (HCD56, BV605), CRTH2 (BM16, PerCP/Cy5.5 and PE), FcεR1 (AER-37, APC), KLRG1 (2F1/KLRG1, BV510), TBET (4B10, BV711) and TCRα/β (IP26, APC); from Invitrogen Scientific, CD14 (61D3, APC), CD16 (CB16, APC), CD11c (3.9, APC), CD127 (RDR5, FITC), CD3 (OKT3, Alexa Fluor 700), CD5 (L17F12, APC) and CD19 (HIB19, AF700).
To detect human LT, single-cell suspensions were incubated with recombinant human LTβR–Fc chimeric protein (2 µg ml−1, R&D systems, 629-LR) in PBS with 1% FBS and 0.5 mM EDTA (pH 8.0, Invitrogen) for 30 min in the dark at 4 °C followed by incubation with a secondary antibody (mouse anti-human IgG, Invitrogen) for 30 min in the dark at 4 °C. All of the samples for flow cytometry were from prospectively collected unselected patients (Supplementary Table 5).
H&E staining
Pancreatic tumours were cut into 2-mm-thick slices and fixed in 4% paraformaldehyde solution (Electron Microscopy Sciences), embedded in paraffin, stained with H&E, and scanned on the Panoramic Scanner (3DHistech) with the ×20/0.8 NA objective. The number of TLSs were determined in at least three sections using QuPath (v.0.2.3; https://qupath.github.io/). A compact aggregate of lymphocytes >5,000 µm2 was considered as a TLS71. In DSS-colitis mice, lymphoid aggregates were counted in a section of a colon and normalized by the length of each colonic section.
Serum IL-33 detection
Human
Serum was collected and stored (−80 °C) until use. IL-33 levels were measured by multiplexed cytokine array (Human Cytokine/Chemokine 71-Plex Discovery Assay Array; Eve Technologies). Patient clinicopathological characteristics are shown in Supplementary Table 3.
Mouse
Serum was collected 48 h after the last rIL-33 dose and stored (−80 °C) until use. IL-33 levels were measured by multiplexed cytokine array (Mouse Cytokine Th17 12-Plex Discovery Assay, Eve Technologies).
Immunohistochemistry
Immunohistochemistry was performed on previously described human PDAC tissue microarrays16. In brief, paraffin-embedded tissue sections were deparaffinized with EZPrep buffer (Ventana Medical Systems). Antigen retrieval was performed with CC1 buffer (Ventana Medical Systems), followed by Background Buster solution (Innovex). Avidin-biotin blocking solution (Ventana Medical Systems) was then used to block tissue sections for 30 min. The sections were incubated with anti-human IL-33 antibody (AF3625, R&D System) for 4 h, followed by 60 min with biotinylated rabbit anti-goat IgG (Vector labs) at 1:200 dilution. IL-33 positivity was detected using the DAB detection kit (Ventana Medical Systems). Any cell demonstrating cytoplasmic or nuclear IL-33 positivity was designated to have positive staining. Nucleated cells in a TLS were determined and counted using the Analyse Particles function in ImageJ (v.2.3.0, NIH). IL-33+ cells in a TLS were counted manually.
Immunofluorescence
Three-colour
Paraffin-embedded tissues were sliced into 7 µm sections. Multiplex immunofluorescence was performed using a Discovery XT processor (Ventana Medical Systems) as described previously29. To stain slides for B220, the sections were incubated with anti-B220 (RA3-6B2, BD Biosciences) for 6 h, followed by 60 min incubation with biotinylated horse anti-goat IgG (Vector Laboratories) at 1:200 dilution. Detection was performed with Streptavidin-HRP D (Ventana Medical Systems), followed by incubation with Tyramide Alexa Fluor 594 (Invitrogen) prepared according to the manufacturer’s instructions with predetermined dilutions. The sections were then incubated with anti-CD3 (Dako) for 6 h, followed by 60 min incubation with biotinylated goat anti-rabbit IgG (Vector Laboratories) at 1:200 dilution. Detection was performed with Streptavidin-HRP D (Ventana Medical Systems), followed by incubation with Tyramide Alexa 488 (Invitrogen) prepared according to the manufacturer’s instructions with predetermined dilutions. Finally, the sections were incubated with anti-LYVE-1 (R&D systems) for 6 h, followed by 60 min incubation with biotinylated goat anti-rabbit IgG (Vector Laboratories) at 1:200 dilution. Detection was performed with Streptavidin-HRP D (Ventana Medical Systems), followed by incubation with Tyramide Alexa 647 (Invitrogen) prepared according to the manufacturer’s instructions with predetermined dilutions. After staining, the slides were counterstained with DAPI (Sigma-Aldrich) for 10 min and cover-slipped with Mowiol.
Five-colour in mice
Paraffin-embedded tissues were sliced into 7 µm sections. Multiplex immunofluorescence was performed using a Discovery XT processor (Ventana Medical Systems) as described29. The sequential antibody staining was conducted as follows: 6 h incubation with anti-MECA-79 (BD Pharmingen) followed by 60 min incubation with secondary antibody (Al488); 6 h incubation with anti-BCL6 (Abcam) followed by 60 min incubation with secondary antibody (CF594); 6 h incubation with anti-CD11c (Cell Signaling) followed by secondary antibody (Al647); 6 h incubation with anti-CD3 (Dako) followed by secondary antibody (CF543); and 6 h incubation with anti-B220 (BD BioScience) followed by secondary antibody (CF430). For TLS quantification, the number of TLSs was determined in at least three sections using CaseViewer (3DHISTECH). Mouse TLSs were determined as a compact aggregate of T and B lymphocytes and high endothelial venules. TLS maturation states72,73,74 were defined as follows: lymphoid aggregates, CD3+B220+CD11c−MECA-79−BCL6−; primary follicles, CD3+B220+CD11c+MECA-79+BCL6−.
Five-colour in humans
Automated multiplex immunofluorescence was conducted using the Leica Bond BX staining system. Paraffin-embedded tissues were sectioned at a thickness of 5 μm and baked at 58 °C for 1 h. The slides were loaded in Leica Bond and staining was performed as follows. The samples were pretreated with EDTA-based epitope retrieval ER2 solution (Leica, AR9640) for 20 min at 100 °C. The 4-plex antibody staining and detection were conducted sequentially. The primary antibodies against IL-33 (1.25 μg ml−1, R&D Systems), MECA-79 (1.25 μg ml−1, BD Pharmingen), CD21 (5 μg ml−1, Leica Biosystems), CD45 (0.625 μg ml−1, Abcam) and CD3 (1:10, Roche Diagnostics) were used.
For rabbit antibodies, Leica Bond Polymer anti-rabbit HRP was used; for goat, rat and mouse antibodies, rabbit anti-goat (Jackson ImmunoResearch), rabbit anti-rat (Vector Laboratories) and rabbit anti-mouse (Abcam) secondary antibodies were used before applying the Leica Bond Polymer anti-rabbit HRP. Next, Alexa Fluor tyramide signal amplification reagents (Life Technologies) or CF dye tyramide conjugates (Biotium) were used for detection. After each round of immunofluorescence staining, epitope retrieval was performed to denature primary and secondary antibodies before applying another primary antibody. The slides were then washed in PBS and incubated in 5 μg ml−1 DAPI (Sigma-Aldrich) in PBS for 5 min, rinsed with PBS and mounted in Mowiol 4–88 (Calbiochem). The slides were kept overnight at −20 °C before imaging. CD45 and IL-33 positivity was determined by CaseViewer (3DHISTECH). IL-33+ and IL-33+ CD45+ cells were quantified on ImageJ (National Institute of Health), and cells per area estimated by calculating surface area based on presence of DAPI+ cells. Human TLSs were determined as compact aggregate structures of T cells (CD45+CD3+)7,75, B cells (CD45+CD21+)7,75 and high endothelial venules (MECA-79+)6.
B cell receptor sequencing
Mouse B cell receptor sequencing was done by Adaptive Biotechnology. In brief, ten 5 µm curls of FFPE blocks for each sample were preserved at −80 °C. DNA was extracted, and immunosequencing of the CDR3 regions of mouse B cell receptor chains was performed on genomic DNA from FFPE-fixed samples using the immunoSEQ Assay (Adaptive Biotechnologies76). All of the samples were amplified in a bias-controlled multiplex PCR, followed by high-throughput sequencing, identification and quantification of absolute abundances of unique CDR3 regions. The resulting sequencing data were processed and analysed on the immunoSEQ Analyser web-based relational database.
Somatic hypermutation
All rearrangements were exported to FASTA files using the Fasta Conversion tool in the Immunoseq Analyzer web-based platform and input to the nf-core/airrflow pipeline for analysis using the Immcantation toolset77. Each rearrangement was annotated with its germline V(D)J gene allele using IgBlast with the Mus musculus IMGT germline reference v.2022.10.04 (http://www.imgt.org/IMGT_GENE-DB). Rearrangements with productive heavy-chain sequences were retained for analysis of B cell clonal relationships, and clones were defined by VDJ-aware spectral clustering using the R package SCOPer78. Mutation frequencies were computed using the observed mutations function in the R package Shazam79.
Digital droplet PCR
For purified cells, frozen cells were lysed in 1 ml of TRIzol Reagent (Thermo Fisher Scientific), and phase separation was induced with 200 µl of chloroform. RNA was extracted from 350 µl of the aqueous phase using the miRNeasy Micro Kit (Qiagen) on the QIAcube Connect (Qiagen) according to the manufacturer’s protocol. The samples were eluted in 30 µl of RNase-free water.
For whole intestine, frozen tissues were homogenized in in TRIzol (Thermo Fisher Scientific), and phase separation was induced with 200 µl of chloroform. RNA was extracted from the aqueous using the MagMAX mirVana Total RNA Isolation Kit (Thermo Fisher Scientific) on the KingFisher Flex Magnetic Particle Processor (Thermo Fisher Scientific) according to the manufacturer’s protocol with 350 µl input. Samples were eluted in 35 µl of elution buffer.
PCR expression probes were designed (Bio-Rad) for the following genes: mouse Il33 (dMmuCPE5096722), Ltbr (dMmuCPE5113608), Ccl19 (dMmuCPE5092188), Cxcl9 (dMmuCPE5122450) and Cxcl13 (dMmuCPE5110356). Droplet generation was performed on the QX200 ddPCR system (Bio-Rad) using cDNA generated from 0.8–2 ng total RNA with the One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad) according to the manufacturer’s protocol with reverse transcription at 42 °C and annealing/extension at 60 °C. Each sample was evaluated in technical duplicates. Plates were read and analysed (QuantaSoft) to assess the number of droplets positive for genes of interest. The number of droplets was normalized to the input amount of total RNA.
scRNA-seq
Library preparation, sequencing and post-processing for single-cell immune profiling have been previously described16. In brief, ST2+ ILC2s were purified from tumours and DLNs from PDAC mice treated with rIL-33 for 10 days, and CD45+CD11b+LTBR+ and CD45+CD11b+LTBR− cells were purified from tumours of PDAC mice treated with rIL-33 for 14 days. scRNA-seq libraries were prepared according to the manufacturer’s recommendations (Chromium Single Cell V(D)J User Guide PN-1000006, 10x Genomics). Cell suspensions (85–90% viable) at a concentration of 90–200 cells per μl were loaded onto the 10x Genomics Chromium platform to generate Gel Beads in Emulsion (GEMs), targeting about 2,000 single cells per sample. After GEM generation, the samples were incubated at 53 °C for 45 min in the C1000 Touch Thermal cycler with a 96-Deep Well Reaction Module (BioRad) to generate poly(A) cDNA barcoded at the 5′ end by the addition of a template switch oligo linked to a cell barcode and unique molecular identifiers (UMIs). After breaking the GEMs, the single-stranded cDNA was cleansed with DynaBeads MyOne Silane Beads (Thermo Fisher Scientific). The cDNA was then amplified (98 °C for 45 s; then 16 cycles of 98 °C for 20 s, 67 °C for 30 s, 72 °C for 1 h), after which the cDNA quality was assessed using the Agilent Bioanalyzer 2100, obtaining a product of about 1,200 bp. cDNA (50 ng) was enzymatically fragmented, end repaired, A-tailed, subjected to a double-sided size selection with SPRI select beads (Beckman Coulter) and ligated to adaptors provided in the kit. Within each library, a unique sample kit was then introduced through 14 cycles of PCR amplification using the indexes provided in the kit (98 °C for 45 s; 14 cycles of 98 °C for 20 s, 54 °C for 30 s, 72 °C for 20 s; 72 °C for 1 min; then held at 4 °C). A second double-sided selection was then performed on the indexed libraries, after which the libraries were quantified using Qubit fluorometric quantification (Thermo Fisher Scientific). The Agilent Bioanalyzer 2100 was used to assess the quality (average library size 450 bp), after which cDNA was amplified with 18 cycles, and a unique sample index was added to each library in 16 cycles. Diluted libraries were then clustered using the NovaSeq 600 system on a paired-end read flow cell, sequenced for 28 cycles on R1 (10x barcode and the UMIs), followed by 8 cycles of 17 index (sample index), and 89 bases on R2 transcript, obtaining approximately 100 million clusters per samples. Primary processing of sequencing images was done using Illumina’s Real Time Analysis software (RTA). 10x Genomics Cell Ranger Single Cell Software suite v.3.0.2 (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cellranger) was used to demultiplex samples, align to the mouse genomic reference mm10, filter, count UMIs, single-cell 5′ end genes and control quality according to the manufacturer’s parameters. Processed data were subsequently analysed in R (v.4.0.3).
To estimate a TLS chemokine signature7 in myeloid cells, a signature score was computed for the 12 chemokines7 using the GSVA method80 in R (v.4.0.3). In brief, to achieve quality control, low-quality cells were filtered out based on the median absolute deviation of unique feature counts and mitochondrial counts81, followed by normalization using a global-scaling method. Assay data were then extracted and applied as the expression set, with the 12 chemokines (CCL2, CCL3, CCL4, CCL5, CCL8, CCL18, CCL19, CCL21A, CCL21B, CXCL9, CXCL10, CXCL11 and CXCL13) provided as a reference gene set using the GSVA function. The ensuing consensus expression was projected onto target cell populations using UMAP and violin plot visualizations.
TLS and ILC2 transcriptional signatures
Known TLS gene signatures were extracted from each transcriptomic dataset (see the ‘Tumour transcriptomic profiling’ section). In brief, the signatures included the genes CD79B, EIF1AY, PTGDS, CCR6, SKAP1, CETP and CD1D in ref. 4; CCL2, CCL3, CCL4, CCL5, CCL8, CCL18, CCL19, CCL21, CXCL9, CXCL10, CXCL11 and CXCL13 in ref. 7; and CXCL13, CD200, FBLN7, ICOS, SGPP2, SH2D1A, TIGIT and PDCD1 in ref. 8. The signature score was calculated as the mean gene expression. Pearson’s correlation tests were performed using the rcorr function of the Hmisc package (v.4.5) in R (v.4.0.3).
To establish a human intratumoural ILC2 transcriptional signature (ILC2 score), we first identified candidate differentially expressed genes in rIL-33-activated KLRG1+ ILC2s in tumours and DLNs of PDAC mice, based on purified single-cell transcriptomes (from scRNA-seq above) using the Seurat package on R. After unsupervised clustering, we computed differentially expressed genes for each cluster in the dataset shown in Fig. 3a using Wilcoxon rank-sum test with adjustment for multiple comparisons. As a result, we identified 341 genes significantly and specifically upregulated in KLRG1+ ILC2s (cluster 0).
We next identified human orthologues of the 341 genes upregulated in mouse KLRG1+ ILC2s using the Ensembl Biomart database82, and identified genes with unique human orthologues. We next downloaded the raw read counts for the primary pancreatic cancers (PAAD) from the RNA-seq TCGA dataset using TCGAbiolinks R package83,84,85. We removed non-coding genes from the read count matrix, as well as genes with <1 read per million in at least 5% of the samples in the dataset. We then restricted the ILC2 gene list to the genes expressed (as defined above) in the TCGA PAAD dataset, ordered them by the statistical significance in the mouse differential expression analysis and selected the top 20 genes to serve as the human ILC2 score (LAPTM5, IL7R, TSPAN13, LY6E, S100A10, S100A6, HSPA8, SELPLG, ITM2B, B2M, CORO1A, PFN1, MYL12B, CNN2, RAC2, TSPO, TMSB4Y, PTPN18, CD52 and RPL37A).
To evaluate the relationship between this ILC2 score and TLS signatures4,7,8, we computed the log-transformed normalized gene expression in the TCGA-PAAD dataset described above using the TMM method86 from edgeR package. Consensus expression of the three existing TLS signatures and the new ILC2 score was computed using the GSVA method80. We ran t-tests for Pearson correlations between the consensus expression of the ILC2 score and each of the three existing TLS signatures and applied the Bonferroni correction for multiple testing.
Confocal microscopy
To visualize LT expression on KLRG1+ ILC2s, PDAC mice were treated with rIL-33 for 2 weeks, intratumoural KLRG1+ ILC2s were sort purified and surface LT expression was detected with recombinant mouse LTβR–Fc chimeric protein (1 µg ml−1, R&D systems) as described above (see the ‘Flow cytometry’ section). Sort-purified KLRG1+ ILC2s and MACS-purified CD3/28-stimulated T cells were incubated with recombinant mouse LTβR–Fc chimeric protein (1 µg ml−1, R&D systems) in 5% FBS at 4 °C for 30 min. Cells were then stained with PE-conjugated anti-mouse IgG2a antibody (Invitrogen) at 4 °C for 30 min. Cells were then settled on 0.01% polylysine-coated coverslips for 30 min at room temperature, fixed on coverslips with 4% PFA and permeabilized with 0.1% Triton X-100 for 20 min at room temperature. The coverslips with fixed cells were mounted with Molecular Probes SlowFade Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific).
To visualize LT–PE and nuclear DAPI, the images were taken on a point laser-scanning confocal system (SP5, Leica) with a ×63 oil-immersion using objective 2 (×2 visual magnification). Images with binomial signals were visualized using ImageJ.
In vitro studies
ST2 reporter assay
A total of 5 × 104 HEK-Blue IL-33 cells (Invivogen) was seeded onto 96-well plates with DMEM, 10% FBS, penicillin (100 IU ml−1) and streptomycin (100 µg ml−1). Cells were incubated for 24 h at 37 °C in 5% CO2 with H-rIL-33 (Proteos), H-e-rIL-33 or H-e-rIL-33–Fc at the designated concentrations. After incubation, 20 µl of supernatant was added to 180 µl of QUANTI-Blue solution (Invivogen) per well in a flat-bottom 96-well plate. The plate was incubated for 2 h at 37 °C in 5% CO2 followed by 630-nm-wavelength absorbance detection on the Cytation 3 reader (BioTek).
Statistics
Comparisons between two groups were performed using unpaired Mann–Whitney U-test with the Benjamini–Krieger–Yekutieli false-discovery approach for multiple-timepoint comparisons (two-tailed). Comparisons among multiple groups were performed using one-way ANOVA followed by Kruskal–Wallis multiple-comparison post-test. Comparisons among multiple groups across multiple timepoints were performed using two-way ANOVA followed by Šidák’s multiple-comparison post-test. EC50 curves were compared using an extra sum of squares F test. Correlations between two variables were calculated using linear regression. All alpha levels were 0.05; P < 0.05 was considered to be a significant difference. Statistical analyses were performed using R (v.4.0.3, scRNA-seq) and Prism v.9.2.0 (GraphPad Software, all else).
Material availability
All material is available from the corresponding author on reasonable request.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.