Cell culture
HEK (HEK Flp-In T-Rex 293, Invitrogen) cells were cultured in DMEM (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Gibco) under standard tissue culture conditions (37â°C, 5% CO2). HEK293F (Thermo Fisher Scientific) cells were cultured in Freestyle medium (Thermo Fisher Scientific) at 37â°C, 8% CO2 and 120ârpm. Cells were negative for mycoplasma contamination.
Native PAGE
For immunoblotting, when HEK cells were at about 80% confluency, they were washed twice with ice-cold PBS and scraped in PBS, pelleted by centrifugation for 5âmin at 1,000g, 4â°C and resuspended in modified native lysis buffer (50âmM HEPES pHâ7.4, 50âmM KCl, 1.5âmM MgCl2, 10% glycerol, 0.1% NP-40, 1âmM PMSF, complete EDTA-free protease inhibitor cocktail and 1 mM DTT). Lysis buffer was also supplemented with 30âUâmlâ1 benzonase to remove DNA. Lysis was performed on ice for 20âmin and the lysates were clarified by centrifugation for 10âmin at 12,000g at 4â°C. The protein concentration was determined using a BCA assay (Thermo Fisher Scientific). 4à NativePAGE sample buffer (Thermo Fisher Scientific) was added to a final concentration of 1Ã. Then, 15âµg of each sample was resolved on 3â12% Bis-Tris NativePAGE gels (Thermo Fisher Scientific). NativePAGE was soaked in 0.1% SDS buffer for 15âmin, then transferred to 0.45âµM PVDF membranes presoaked in methanol for 30âs. The membranes were blocked with 5% molecular biology grade BSA (Millipore Sigma) in Tris-buffered saline supplemented with 0.1% Tween-20 (TBST) for 1âh at room temperature, then probed with specific primary antibodies 4â°C for overnight. Primary antibodies was diluted in 1% BSA/TBST as follows: 1:10,000 rabbit anti-CCT5 (Abcam, ab129016). The secondary antibody was diluted 1:10,000 in TBST. Total protein was detected with Revert total protein stain. Fluorescence signal detection was performed using Li-Cor Odyssey infrared imager.
PDCD5 knockdown
HEK cells (5âÃâ105) were seeded into six-well plates. Then, 24âh after plating, 25âpmol siRNA (Thermo Fisher Scientific, s17467) were added with Lipofectamine RNAiMAX Transfection Reagent (Invitrogen). Cells were collected with ice-cold PBS after 48âh and then immunoblotting was run for further analysis.
Expression and purification of recombinant PDCD5 and its mutants
PDCD5 mutants were obtained using site-directed mutagenesis. A 6Ã His-tag was added to the C terminus of PDCD5. Plasmids containing WT and mutant PDCD5 were transformed into Escherichia coli Rosetta DE3 competent cells for expression. PDCD5 was expressed and purified as previously reported33. In brief, cell lysates were first passed through a nickel column, then PDCD5 bound to the nickel resin was eluted in high imidazole buffer, and pure PDCD5 was obtained by passing the elution twice through a Superdex 200 size-exclusion column. Proteins were concentrated by centrifugation and then quantified using the BCA colorimetric assay.
TRiC ATPase activity
The assay was performed as previously described48. In brief, stock solutions of 0.05% (w/v) quinaldine red, 2.32% (w/v) polyvinyl alcohol, 5.72% (w/v) ammonium heptamolybdate tetrahydrate in 6âM HCl and water were mixed in a 2:1:1:2 ratio to prepare the quinaldine red reagent fresh before each experiment. Then, 300ânM TRiC was diluted in ATPase buffer (50âmM Tris-HCl pHâ7.4, 100âmM KCl, 5âmM MgCl2, 10% glycerol, 1âmM TCEP; 30âμl total reaction volume), preheated to 37â°C and added to 3âμl water or 10âmM ATP to start the reaction, then incubated for the indicated durations in the presence or absence of 3âµM PDCD5. The reactions were stopped by the addition of 5âμl of 60âmM EDTA in a Corning 96-well opaque non-sterile polystyrene plate (Sigma-Aldrich, CLS3992) on ice. After samples at all timepoints were collected, the reactions were developed by adding 80âμl quinaldine red reagent for 10âmin, then quenched by adding 10âμl 32% (w/v) sodium citrate. The fluorescence intensity was measured (excitation, 430ânm; emission, 530ânm) using the CLARIOstar plate reader (BMG Labtech). Analysis was performed by fitting a phosphate standard curve with a one-phase decay function, and we derived the parameters for calculating the amount of phosphate released from CCT complexes.
PDCD5 binding to TRiC
To probe the binding affinity of PDCD5 for TRiC, increasing amounts of recombinant PDCD5 variants were incubated with a fixed concentration of TRiC (300ânM) for 20âmin at 25â°C in ATPase buffer (50âmM Tris-HCl pHâ7.4, 100âmM KCl, 5âmM MgCl2, 10% glycerol, 1âmM TCEP), in the absence of ATP. The reactions were run in native gels and immunoblotted using PDCD5 or CCT8 antibodies, as described above. To test whether PDCD5 binds to the TRiC open or closed conformations, 3âμM of WT or mutant PDCD5 was incubated with 300ânM TRiC for 20âmin at 25â°C in ATPase buffer containing 1âmM of different nucleotides and ATP analogues. The reactions were run in native gels and immunoblotted using PDCD5 or CCT8 antibodies, as described above. To obtain insights about the binding kinetics of PDCD5 variants to TRiC, 3âμM of WT or mutant PDCD5 was incubated with 300ânM TRiC in ATPase buffer at 25â°C for 10, 15, 20 and 30âmin. The reactions were run in native gels and immunoblotted using PDCD5 (Proteintech, 12456-1-AP, 1:1,000) and CCT8 (Santa Cruz Biotechnology, sc-377261, 1:250) antibodies, as described above.
Co-IP
For PDCD5âFlag co-IP, PDCD5-Flag constructs (GenScript) were transiently expressed in HEK293F for 48âh after transfection. Cells were washed with PBS before collection by centrifugation and frozen in liquid nitrogen. HEK293F cells were lysed in lysis buffer (PBS pHâ7.4, 0.1% IGEPAL CA-630, 5âmM MgCl2, freshly added 0.6âmM phenylmethylsulphonyl fluoride and protease inhibitors), triturated through a 24-gauge needle ten times and incubated on ice for 5âmin. After lysate clearing by centrifugation, 500âμg clarified protein extract was mixed with 20âµl packed anti-Flag M2 beads (Sigma-Aldrich) and incubated for 1âh at 4â°C. After three washes with lysis buffer, bound proteins were eluted by boiling in LDS sample buffer (Invitrogen). For western blotting, input and eluate (IP) samples were loaded onto 4â12% Bis-Tris gels (Invitrogen) and subsequently transferred to nitrocellulose membranes (Bio-Rad).
CCT3 co-IP was performed with non-transfected HEK293F cells subjected to in vivo cross-linking with 1.5âmM dithiobis(succinimidyl propionate) (DSP; Thermo Fisher Scientific) at 37â°C for 10âmin. The cross-linking reaction was quenched by the addition of Tris (pHâ8.0) to a final concentration of 160âmM and cells were collected and lysed as described above. Then, 2âmg of clarified protein extract was mixed with 10âμg rabbit anti-CCT3 antibody (Proteintech, 10571-1-AP) or rabbit control IgG (Proteintech, 30000-0-AP) as mock IP for 1âh at 4â°C, followed by addition of 50ââμl equilibrated Protein G Magnetic Beads (Thermo Fisher Scientific) and incubation for 1âh at 4â°C. The samples were washed, eluted and evaluated using SDSâPAGE as described above.
The percentage of IP efficiency was calculated by normalizing the measured intensities and the respective dilution factor of the loaded sample for western blotting (1% for the input sample and 5% for the IP sample), followed by IP/input. For the quantification, the mean ± s.d. values were as follows: PDCD5âflag (42.70â±â16.16), CCT1 (86.66â±â41.01), CCT2 (45.54â±â15.25), CCT3 (45.57â±â12.47), CCT4 (61.12â±â15.08), CCT5 (98.98â±â27.74), CCT6 (53.74â±â21.34), CCT7 (65.99â±â38.51), CCT8 (135.49â±â64.48) and GAPDH (0.03â±â0.06), with n representing the number of biologically independent experiments (nâ=â4). For the quantification of PDCD5 mutation experiments, the meanâ±âs.d. values were as follows: WT (100â±â0), RKK (133.65â±â59.63) and IL (11.04â±â9.68), with n representing the number of biologically independent experiments (nâ=â4).
To induce TRiC closure during co-IP, beads bound with TRiCâPDCD5âFlag (from co-IP, see above) were incubated in ATP/AlFx buffer (lysis buffer supplemented with 5âmM Al(NO3)3, 30âmM NaF and 1âmM ATP) for 1âh at 37â°C, followed by three washes with ATP/AlFx buffer. As a control, the beads bound with TRiCâPDCD5âFlag (from co-IP, see above) were incubated and washed in lysis buffer without the ATP/AlFx. For western blotting, 1% of input, 25% of released proteins after ATP/AlFx incubation and 25% of eluates (denoted as beads) were loaded.
Without adding ATP in the TRiC sample before plunge freezing, around 100% TRiC particles are at open conformation based on the single-particle analysis13,14,19. With extra ATP/AlFx in TRiC solution before plunge freezing, a portion of TRiC particles were closed, although different papers show different closed/open ratios with ATP/AlFx at different conditions. Closed/open ratio: ~1.7 in buffer (1âmM ATP, 5âmM MgCl2, 5âmM Al (NO3)3 and 30âmM NaF) from ref. 13; ~5.1 in buffer (1âmM ATP, 1âmM Al3(NO3)3, 6âmM NaF, 10âmM MgCl2 50âmM KCl) from ref. 21; ~0.6 in buffer with ATP-AlFx from ref. 14; and ~2.2 in buffer (1âmM ATP, 5âmM MgCl2 and AlFx (5âmM Al(NO3)3 and 30âmM NaF) from ref. 16. In our experimental settings (Extended Data Fig. 7), we used the conditions from ref. 13 (1âmM ATP, 5âmM MgCl2, 5âmM Al (NO3)3 and 30âmM NaF).
For the quantification in Extended Data Fig. 7, the meanâ±âs.d. values were as follows: PDCD5 (ATP/AlFx) (0.09â±â0.05); PDCD5 (control) (0.10â±â0.04); CCT1 (ATP/AlFx) (1.53â±â0.51); and CCT1 (control) (0.38â±â0.06); with n representing the number of biologically independent experiments (nâ=â4).
Thermal protein profiling (heat-shock treatment of cells)
WT (Abcam, ab255449) and PDCD5-knockout HEK293T cells (Abcam, ab266229) were used for the heart-shock assay and cultured in DMEM (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Gibco) at 37â°C with 5% CO2. The experiment was conducted as described previously49,50. In brief, cells were collected and resuspended in PBS. Five aliquots were prepared and distributed into PCR tubes, each of the tubes containing 5âÃâ105 cells. Each tube was incubated for 3âmin at various temperatures (37.0, 44.1, 49.9, 55.5 and 62.0â°C; or 56.8, 58.3, 59.5, 60.7 and 62.1â°C). The cells were then lysed in a buffer containing 1.5âMm MgCl2, 0.8% NP-40, 0.4Uâμlâ1 benzonase and protease inhibitor for 40âmin at 4â°C. Protein aggregations were removed, and the soluble fraction was used for western blotting. For quantification of the western blotting of thermal protein profiling, the meanâ±âs.d. values of actin in WT cells at 37.0â°C to 62.0â°C were as follows: 100.0â±â0.0, 85.3â±â5.2, 73.8â±â7.7, 46.3â±â2.9 and 26.3â±â9.4; the meanâ±âs.d. values of actin in PDCD5-knockout cells at 37.0â°C to 62.0â°C were as follows: 100.0â±â0.0, 100.3â±â7.0, 109.0â±â9.7, 83.0â±â2.0 and 57.6â±â9.4; the meanâ±âs.d. values of tubulin in WT cells at 56.8â°C to 62.1â°C were as follows: 100.0â±â0.0, 78.2â±â4.2, 49.3â±â5.5, 20.0â±â4.9 and 5.4â±â3.8; and the meanâ±âs.d. values of tubulin in PDCD5-knockout cells at 56.8â°C to 62.1â°C were as follows: 138.0â±â22.3, 99.7â±â6.4, 63.9â±â15.9, 34.8â±â0.4 and 8.3â±â4.7.
Antibodies
Membranes from western blotting were incubated with primary antibodies (mouse anti-Flag M2 (Sigma-Aldrich, F1804, 1:2,000), rabbit anti-PDCD5 (Abcam, ab126213, 1:1,000), rabbit anti-CCT1 (Abcam, ab240903, 1:10,000), rabbit anti-CCT2 (Abcam, ab92746, 1:10,000), rabbit anti-CCT3 (Proteintech, 10571-1-AP, 1:30,000), rabbit anti-CCT4 (Proteintech, 21524-1-AP, 1:5,000), rabbit anti-CCT5 (Proteintech, 11603-1-AP, 1:3,000), rabbit anti-CCT6 (Proteintech, 19793-1-AP, 1:1,000), rabbit anti-CCT7 (Abcam, ab240566, 1:30,000), rabbit anti-CCT8 (Proteintech, 12263-1-AP, 1:2,000), rabbit anti-GAPDH (Proteintech, 10494-1-AP, 1:15,000), mouse anti-actin (Invitrogen, AM4302, 1:3,000), mouse anti-tubulin (Sigma-Aldrich, T5168, 1:3,000)), followed by incubation with HRP-conjugated secondary antibodies (anti-rabbit IgG (Cell Signaling, 7074, 1:10,000), anti-mouse IgG + IgM (Jackson ImmunoResearch, 115-035-044, 1:10,000)). Uncropped western blots are provided as Source Data.
Grid preparation, data acquisition and tomogram reconstruction
Cryo-ET sample preparation, data collection and tomogram reconstruction were performed essentially as described previously22. In brief, R2/2 gold grids with 200 mesh (Quantifoil) were glow discharged for 90âs and were positioned in 3.5âcm cell culture dishes (MatTek). Then, 2âml HEK Flp-In T-Rex 293 cell suspension, with a concentration of 175,000 cells per ml, was added to the dish. For untreated samples, cells were cultured for 5âh before plunge-freezing. For HHT-treated samples, cells were cultured without HHT for 3âh and subsequently exposed to HHT (Santa Cruz Biotechnology) at a final concentration of 100âµM for 2âh before the plunge-freezing process. The grids were blotted from the backside for 6âs using the Leica EM GP2 plunger under 70% humidity and 37â°C. The grids were rapidly plunged into liquid ethane and stored in liquid nitrogen. Grids were FIB-milled using Aquilos FIB-SEM (Thermo Fisher Scientific). The samples were sputter-coated with an organometallic protective platinum layer using the gas injection system for 15âs. Lamella preparation was performed through a stepwise milling process with gallium ion-beam currents decreasing from 0.5ânA to 30âpA.
The data acquisition area was focused on the cytoplasmic region within the cell. Tilt series were acquired on a Titan Krios G4 (Thermo Fisher Scientific) operated at 300âkV, and equipped with Selectris X imaging filter and Falcon 4 direct electron detector, at 4,000âÃâ4,000 pixel dimensions, pixel size of 1.188âà , a total dose of 120 to 150âeâà â2 per tilt series, 2° tilt increment, tilt range of â60° to 60° and target defocus of â1.5 to â4.5âµm, using SerialEM software51. Tilt series were aligned automatically using the IMOD package52. The alignment files generated from IMOD were used for tomogram reconstruction in Warp53 v.1.0.9.
Particle localization and refinement
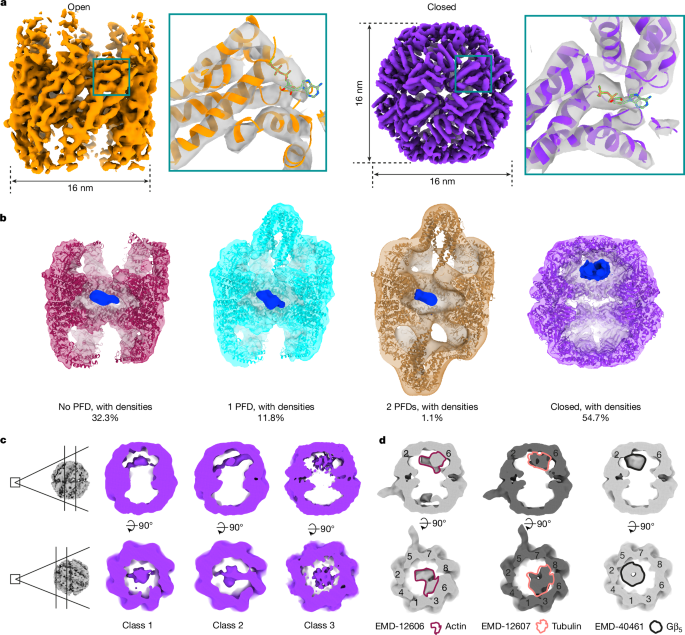
Template matching was performed similarly to previous studies22,54. For this work, the parameters were set as follows: 5° angular scanning step, low-pass filter radius=20, high-pass filter radius=1, apply_laplacian=0, noise_corelation=1 and calc_ctf=1. The cryo-EM map (EMD-32822)14 of TRiC downloaded from the Electron Microscopy Data Bank (EMDB) was used as the template covered by a sphere mask. The above optimized setting produced distinguished peaks visualized in napari55 (Extended Data Fig. 1b and Supplementary Video 1). To analyse all potential TRiC complexes within the datasets, we extracted the top 1,000 peaks per tomogram. The selection was based on the constrained cross-correlation (CCC) value from template matching, and these chosen coordinates were subsequently extracted as subtomograms in Warp. In total, 360,000 untreated and 352,000 treated subtomograms were extracted. 3D classifications (classes = 4, T = 0.5, iterations = 30, without mask) and refinements (C1 symmetry) were performed in RELION56 v.3.1. In total, 3,353 open TRiC particles and 4,054 closed TRiC particles in the untreated dataset, and 3,785 and 3,418 in the treated dataset were identified. Open TRiC particles from untreated and treated datasets were combined and refined to improve map resolution. Closed TRiC particles were merged from untreated and treated datasets and refined with C1 or D8 symmetry. Actin filaments were manually picked in ten tomograms. In total, 1,490 subtomograms were extracted and refined at bin4. Atomic models obtained from the PDB (7X3J, 7NVN, 7NVO, 7NVL, 7NVM and 8F8P)13,16,57 were fitted into our maps. ChimeraX58,59 was used to visualize EM maps and models.
Subtomogram classification of TRiC states
For 3,353 open TRiC particles in the untreated dataset, classification with a sphere mask covering the potential PFD region (classes = 3, T = 3, iterations = 50, C1 symmetry) of one ring (denoted ring1) was performed (Extended Data Fig. 2a), which generated 2,874 particles without PFD and 479 particles with PFD of ring1. Independently, the same classification was performed with a mask focused on the other ring (denoted ring2), which produced 2,791 particles without PFD and 562 particles with PFD of ring2. In total, 2,395 particles without PFD, 875 particles with 1 PFD and 83 particles with 2 PFD were identified by sorting particles based on the above two classifications. The same classification strategy was applied to 3,785 open TRiC particles in the treated dataset, resulting in 2,334 particles without PFD, 1,287 particles with 1 PFD and 164 particles with 2 PFD. The atomic model (PDB: 7WU7)14 was fitted into the maps with PFD. Different classification parameters were evaluated in attempts to resolve the density in the chamber of TRiC, but this did not result in meaningful insights. The densities inside the TRiC chamber were Gaussian filtered (sDevâ=â2 or 4) for visualization in Figs. 1b and 4d and Extended Data Figs. 3 and 10. For closed TRiC, 3D classification (classes = 4, T = 3, iterations = 35, C1 symmetry) was performed in untreated and treated datasets independently in RELION 3.1, which revealed several classes with different densities occupied in the chamber of the closed TRiC. Further classification with a mask focusing on the substrate position did not produce meaningful results (Supplementary Figs. 4 and 5). Fourier shell correlation (FSC) was calculated in RELION 3.1.
AlphaFold-Multimer model of the CCT3âCCT1âCCT4âPDCD5 complex
The structure of human PDCD5 in a complex with human CCT3, CCT1 and CCT4 was predicted using AlphaFold-Multimer31 (v.2.2.0). The prediction was executed using the default setting with AMBER relaxation, and 15 models were generated for each prediction. The same prediction setting was used for PDCD5 with the other CCT combinations. The full-length amino acid sequences of PDCD5 (UniProt: O14737)60 and the equatorial domain of CCT1âCCT8 (the sequences were the same as PDB 7NVO) were used for the above prediction. The monomeric model of PDCD5 (AF-O14737-F1) was downloaded from the AlphaFold Protein Structure Database30.
Sequence alignment
Sequence alignment of CCT1âCCT8 (UniProt: P17987, P78371, P49368, P50991, P48643, P40227, Q99832 and P50990) was executed through Clustal Omega61. Sequence alignment of PDCD5 (UniProt: M. maripaludis, A9A8D7; S. pombe, O13929; C. elegans, Q93408; mouse, P56812; bovine, Q2HJH9; and human, O14737) and CCT1 (UniProt: H. volcanii, O30561; S. pombe, O94501; C. elegans, P41988; mouse, P11983; bovine, Q32L40; and human, P17987) were performed with ClustalO in Jalview62. The sequence conservation score of PDCD5 was calculated using the ConSurf server63.
Spatial analysis of TRiC in situ
The distance and angle examination of TRiC was performed similarly to as in previous studies22,64,65. For TRiC cluster tracing, the coordinates of TRiC determined by subtomogram averaging were used to localize the particles in the tomograms. The TRiC cluster (containing â¥2 TRiC particles) was defined by the distance between the coordinates of one TRiC and that of its nearest neighbour using a distance cut-off of 20ânm (centre-to-centre distance). As the coordinate represents the centre of the structure, the rotation of the particles would not affect the distance measurement. The particle closest to the previous particle in terms of Euclidean distance was selected as the trailing TRiC within the cluster, provided that it fell within the permitted distance threshold. Various distance thresholds ranging from 15ânm (the minimum centre-to-centre distance between two TRiC) to 40ânm were investigated (Fig. 4b,c). For each specific distance, the threshold was confined within a range of ±0.5 nm (for example, for 17ânm, the permissible distance ranged from 16.5ânm to 17.5ânm). A distance threshold of 20ânm was used to define whether TRiC belongs to the same cluster in this study.
For the distance of TRiC pair analysis in Extended Data Fig. 9h,i, the number and the meanâ±âs.d. values were n2 (cluster length = 2)â=â326 (17.35â±â1.18); n3â=â218 (17.44â±â1.27), n4â=â74 (17.01â±â1.17), n5â=â35 (17.05â±â1.16), n6â=â16 (16.87â±â1.01) and n7â=â4 (17.33â±â0.89), respectively, in the untreated dataset. The number and the meanâ±âs.d. were n2â=â195 (17.04â±â1.28), n3â=â116 (17.42â±â1.18), n4â=â27 (16.87â±â0.96), n5â=â7 (17.09â±â1.25) and n6â=â4 (16.65â±â1.88), respectively, in the treated dataset. TRiC pairs with distances between 15 and 20ânm were analysed.
The angle between TRiC and its closest neighbouring TRiC was investigated for particles within clusters in the untreated dataset (Extended Data Fig. 8d). The divided area of the hemisphere contains all points denoting cone rotation, described by Euler angles θ and Ï, of a vector (0, 0, 1). These rotations are projected onto the northern hemisphere (for vectors rotated with a z-coordinate greater than 0) and the southern hemisphere (for vectors rotated with a z-coordinate less than or equal to 0) using stereographic projection. The north pole corresponds to zero rotation, signifying a vector (0, 0, 1). The rotations of the neighbour TRiC were multiplied by the inverse rotations of the respective neighbour particles.
To calculate the percentage of TRiC clusters with neighbouring actin filaments. The particles from the subtomogram averaging of TRiC and actin filaments were mapped back to tomograms for analysis. The threshold of the neighbouring distance (TRiC centre to the centre of actin dimer) was set to 20ânm.
Spatial relation between ribosomes and TRiC in cells
The spatial distribution of TRiC near the ribosome exit tunnel was investigated. The coordinates of ribosome, 60S and 40S determined by subtomogram averaging were used to localize the particles in the tomograms22. The ribosome was rotated to a reference position (zero rotation) through an inverse rotation, which means it was rotated by (âÏ,ââθ,ââÏ)ribosome. Subsequently, TRiC underwent rotation by its respective angles (Ï,âθ,âÏ)TRiC, followed by another rotation of (âÏ,ââθ,ââÏ)ribosome, therefore aligning the ribosomeâTRiC within a standard rotation frame (zero rotation of the ribosome), while maintaining their original angular relationship. The coordinates of the ribosome exit tunnel were subtracted from both the ribosome exit tunnel coordinates (setting it to zero) and TRiC coordinates. The new TRiC coordinates were rotated by (âÏ,ââθ,ââÏ)ribosome to illustrate their positioning relative to the zero rotation of the ribosome. For the spatial analysis of ribosome and TRiC, ribosome particles were more abundant than TRiC particles. As a result, the same TRiC can be the nearest neighbour of several ribosomes. Our analysis focused on the ribosomes that acted as the nearest neighbours of TRiC. The meanâ±âs.d. in Extended Data Fig. 9c,k were as follows: untreated open TRiC in the ribosome ETS (55.1â±â0.8%); untreated closed TRiC in the ETS (55.3â±â0.3%); untreated open TRiC in the non-ETS (44.9â±â0.8%); untreated closed TRiC in the non-ETS (44.7â±â0.3%); treated open TRiC in the ETS (50.4â±â0.4%); treated closed TRiC in the ETS (49.7â±â1.0%); treated open TRiC in the non-ETS (49.6â±â0.4%); and treated closed TRiC in the non-ETS (50.3â±â1.0%). Data plotting and statistical analysis were performed using GraphPad Prism (v.10, GraphPad Software).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.