Cohort descriptions and phenotypes
DDD
The aim of the DDD study is to find molecular diagnoses for families and patients affected by previously genetically undiagnosed, severe developmental conditions. Recruitment was conducted from 2011 to 2015 across 24 clinical genetics services in the United Kingdom and Ireland58. The clinical inclusion criteria included neurodevelopmental conditions, congenital, growth or behavioural abnormalities and dysmorphic features. Probands were systematically phenotyped through DECIPHER59 using Human Phenotype Ontology (HPO)60 terms and a bespoke online questionnaire that collected information on developmental milestones, growth measurements, number of affected relatives, prematurity, maternal diabetes, and other clinically relevant parameters. The cohort has been described extensively1,50,58,61.
We focused on probands in the DDD cohort who had neurodevelopmental conditions, which were defined previously by Niemi et al.2 Briefly, these were probands who had at least one of the following neurodevelopmental HPO terms or their descendent terms: abnormality of higher mental function (HP:0011446), neurodevelopmental abnormality (HP:0012759), abnormality of the nervous system morphology (HP:0012639), behavioural abnormality (HP:0000708), seizures (HP:0001250), encephalopathy (HP:001298), abnormal synaptic transmission (HP:0012535), or abnormal nervous system electrophysiology (HP:0001311).
GEL project
The 100,000 Genomes project is an initiative by the UK Department of Health and Social Care to sequence the whole genomes of individuals with rare conditions or cancer in the National Health Service62,63. The rare disease branch of the project consists of sequencing data from roughly 72,000 patients with rare conditions and their relatives, in roughly 34,000 families with a variety of structures. There are more than 190 rare conditions represented in the cohort, and about 23% of the patients have neurodevelopmental conditions. The cohort was sequenced at around 35 times coverage, and variant calling and quality control (QC) were performed by Genomics England63,64.
Patients from GEL with neurodevelopmental conditions were defined as those recruited under the âNeurodevelopmental disordersâ disease subcategory, or with more than one HPO term that was a descendant of âNeurodevelopmental Abnormalityâ (HP:0012759). We removed probands whose age of onset was above 16âyears or who had neurodegenerative conditions.
The set of unrelated GEL controls included patients with cancer above 30âyears old (Nâ=â10,469) and unaffected relatives (Nâ=â3,198) of probands with rare conditions who were not in the neurodevelopmental condition set and did not have phenotypes similar to probands from DDD (âDDD-likeâ). The DDD-like probands were defined as those who:
-
1.
were recruited into a disease model that was also used to recruit probands who had previously been recruited into DDD (section below on identifying probands overlapping between the two cohorts), or
-
2.
had one the top five HPO terms used in DDD and their descendants, namely HP:0000729 (autistic behaviour), HP:0001250 (seizure), HP:0000252 (microcephaly), HP:0000750 (delayed speech and language development), and HP:0001263 (global developmental delay).
Probands recruited into the neurodegenerative disorders subcategory or with an age of onset greater than 16âyears were removed from the DDD-like set, as were probands recruited into a disease subcategory for which the average age of probands was older than 16âyears.
To define relatedness, we used a file generated by GEL consisting of a pairwise kinship matrix produced using the PLINK2 (refs. 65,66) implementation of the KING robust algorithm67 and a –king-cutoff of 0.0442 (that is, 1/24.5).
Control cohorts
The UK Household Longitudinal Study (UKHLS) cohort consists of a continuation of the British Household Panel Survey of individuals living in the United Kingdom68,69. The Avon Longitudinal Study of Parents and Children (ALSPAC) is a birth cohort study of children born in Avon, England with expected dates of delivery between 1 April 1991 and 31 December 1992 (ref. 70). Eligible pregnant women (Nâ=â13,761) were recruited and their children have been phenotyped extensively over the past 30âyears. Please note that the study website (http://www.bristol.ac.uk/alspac/researchers/our-data/) contains details of all the data that are available through a fully searchable data dictionary and variable search tool. The MCS is a birth cohort study of children born across the UK during 2000 and 2001 from 18,552 families71,72. Further information about recruitment of these cohorts is given in Supplementary Note 4.
Ethics
The DDD study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South Research Ethics Committee and GEN/284/12, granted by the Republic of Ireland Research Ethics Committee). The 100,000 Genomes project was approved by the East of EnglandâCambridge Central Research Ethics Committee (REF 20/EE/0035). Ethical approval for ALSPAC was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees. Ethical approval for each sweep of MCS was obtained from NHS Research Ethics Committees (MREC). Ethical approval for the sixth MCS sweep, which included the collection of saliva samples from children and biological resident parents, was obtained from London-Central REC (MREC; 13/LO/1786).
Preparation of genetic data
Individuals from DDD, UKHLS, ALSPAC and MCS were genotyped on various arrays, whereas GEL individuals were whole-genome sequenced. The available data are summarized here briefly:
A subset of the DDD cohort (all children and several thousand parents) was genotyped on three genotype array chips: the Illumina HumanCoreExome chip (CoreExome), the Illumina OmniChipExpress (OmniChip) and the Illumina Infinium Global Screening Array (GSA). Some probands were genotyped on more than one chip, as shown in Supplementary Fig. 9. In downstream analysis, we used the CoreExome and OmniChip data for analyses of probands, and the GSA and OmniChip data for analyses of trios. QC of CoreExome (including DDD patients and 9,270 UKHLS controls genotyped on the same chip) and OmniChip data were performed by Niemi et al.2 and we performed QC in the GSA data specifically for this paper (Supplementary Tables 13 and 14). The DDD cohort was also exome sequenced, and those data were used for the analyses involving rare variants.
GEL individuals were whole-genome sequenced with 150âbp paired-end reads using Illumina HiSeqX. Variant calling and QC were performed by Genomics England. We used 78,195 post-QC germline genomes from the Aggregated Variant Calls (aggV2) prepared by the GEL team. We kept variants that passed the QC filters shown in Supplementary Table 15.
Data we received from ALSPAC were processed in two batches69. In the first batch, we received post-QC array data for G0 mothers (Nâ=â8,884) who were genotyped on the Illumina Human 660W chip and G1 children (Nâ=â8,932) genotyped on the HumanHap550 quad chip. In the second batch, we received another 2,198 parents (G0 mothers and G0 partners73) who were genotyped on the CoreExome array.
We received data for 21,181 MCS samples who were genotyped using the GSA array chip74.
We applied standard QC filters in each dataset separately, described further in Supplementary Methods. We used the maximum subset of unrelated individuals that passed QC. We did not use any statistical methods to predetermine sample sizes.
Genetically predicted ancestry
To avoid spurious results due to population stratification, all genetic analyses were conducted in a genetically homogeneous subset of individuals with genetic similarity to British individuals from the 1,000 Genomes Project75, henceforth referred to as having GBR ancestry. The Supplementary Methods provide detailed information on ancestry inference, but we summarize it briefly here. The identification of GBR-ancestry samples from the DDD CoreExome and OmniChip data was described previously2. To identify individuals of genetically inferred GBR ancestry in DDD GSA samples, we first projected post-QC samples onto 1,000 Genomes phase 3 individuals75 (Supplementary Fig. 10). We then performed another principal component analysis (PCA) within the loosely defined European ancestry subset and identified a homogeneous subgroup (Supplementary Fig. 11) using uniform manifold approximation and projection (UMAP)76. As we merged parentâoffspring trios genotyped on GSA and OmniChip array chips in downstream analysis, we kept GSA individuals who were similar to OmniChip individuals in terms of genetic ancestry in PCA space (Supplementary Fig. 12). In GEL, we used individuals with genetically inferred European ancestry, which were identified by the GEL bioinformatics team. We further restricted to a homogeneous subset (Nâ=â56,249) that represents White British individuals (Supplementary Fig. 13). Array data received from the ALSPAC all had genetically predicted European ancestry, so we did not perform any filtering based on genetic ancestry. We performed similar PCA and UMAP clustering to identify individuals of GBR ancestry in MCS (Supplementary Figs. 14 and 15), and further filtered to individuals who self-reported as being of White ethnicity.
Relatives within and across cohorts
Within each dataset, we identified up to third-degree relatives (kinship coefficient greater than 0.0442 by KING v.2.2.4 (ref. 67) using post-QC genotyped array data or WGS data. We always used a subset of unrelated individuals (that is, more distant than third-degree relatives) in downstream analysis. In analyses using trios, we made sure probands in trios were unrelated and parents were unrelated with parents from other families.
In analyses combining DDD and GEL, we removed from GEL any participants who were also recruited into DDD and/or who were related to DDD participants, and also removed Scottish samples from DDD as we were unable to check whether GEL samples were related to them (Supplementary Methods). We removed individuals from the two birth cohorts who were related to each other or to DDD participants, which left 1,434 and 2,498 trios from ALSPAC and MCS, respectively (Supplementary Methods).
Imputation and post-imputation QC
Imputation of array data was performed in each genotyped cohort separately using the maximum number of variants available after QC. Before imputation, we removed palindromic SNPs, SNPs that were not in the imputation reference panel, and SNPs with mismatched alleles. DDD samples and UKHLS controls who were genotyped on the CoreExome array were imputed with the HRC r1.1 reference panel by Niemi et al.2 DDD GSA and OmniChip samples and ALSPAC samples were imputed to the TOPMed r2 reference panel using the TOPMed imputation server, and the MCS samples to the HRC r1.1 reference panel77,78,79. We kept well-imputed common variants with Minimac4 R2â>â0.8 and MAFâ>â1%. For polygenic score analyses, we subsequently restricted to common variants that passed these QC filters in all genotyped cohorts and also passed QC in the GEL WGS data.
Extraction and QC of rare variants
QC of DDD exome sequencing data and extraction of rare single-nucleotide variants, and insertion and deletions (indels) is summarized in Supplementary Table 16. Indels in the same gene and sample were removed (4% of indels with MAFâ<â1%), as these were often part of complex mutational events that would require haplotype-aware annotation.
For GEL, details of the QC of single-nucleotide variants and indels in the WGS data are provided by the GEL team63,64 and variant QC is summarized in Supplementary Table 15. We use a custom python script to extract rare variants from GEL aggregated WGS variant call format files (aggV2). We filtered genotypes to those with genotype quality (GQ)ââ¥â20 and read depth (DP)ââ¥â10. We removed heterozygous genotypes that did not pass a binomial test of balanced REF and ALT alleles (Pâ<â1âÃâ10â3) or for which ALT/(REFâ+âALT) (AB ratio) was not between 0.2 and 0.8. We further removed variants with missing high-quality genotypes in more than 5% of all samples in aggV2. We removed indels in the same gene and sample for the same reason described above for DDD.
For MCS, details of the QC of exome sequencing data are in Supplementary Methods.
Defining monogenic diagnoses in patients
DDD
The DDD study identified clinically relevant rare variants from exome sequencing and microarray data using a filtering procedure described in ref. 58. The procedure focuses on identifying rare damaging variants that fit an appropriate inheritance mode in a set of genes that cause developmental disorders (DDG2P, https://www.deciphergenomics.org/ddd/ddgenes). Variants that pass clinical filtering are uploaded to DECIPHER59, where the patientsâ clinicians are asked to classify them as definitely pathogenic, likely pathogenic, uncertain, likely benign or benign. We defined âdiagnosedâ probands as those with one or more variants either annotated as pathogenic or likely pathogenic in DECIPHER by their referring clinician, or predicted as pathogenic or likely pathogenic using diagnoses autocoded following the American College of Medical Genetics and Genomics guidelines as described in ref. 1. All remaining probands were classed as âundiagnosedâ. Probands with a de novo diagnosis are those with a de novo mutation in a monoallelic or X-linked DDG2P gene that was either annotated or predicted as pathogenic or likely pathogenic.
GEL
The probands assigned diagnostic status were those included in the Genomic Medicine Service exit questionnaire, in which a clinician evaluated the pathogenicity of variants of interest identified through GELâs custom pipeline. We defined diagnosed probands as those that had a pathogenic or likely pathogenic variant that is annotated as partially or fully explaining their phenotype in this exit questionnaire. Probands with a de novo diagnosis are those whose pathogenic or likely pathogenic variants from the exit questionnaire were annotated as de novo protein-truncating or missense variants in DDG2P monoallelic or X-linked genes. We defined undiagnosed probands as those that were present in the exit questionnaire but not annotated as having a pathogenic or likely pathogenic variant and not annotated as âyesâ or âpartiallyâ in the âcase_solved_familyâ column. We further removed from this undiagnosed set any probands who have potential diagnoses in the Diagnostic Discovery data in GEL, which is a list of variants submitted by researchers that are thought probably to be pathogenic by the GEL clinical team.
Defining trio sample sets in DDD and GEL
The procedure used for filtering trios used in DDD and GEL is shown in Supplementary Fig. 16. Briefly, in DDD, we combined data across GSA and OmniChip arrays and kept trios in which all three members had GBR ancestry and the proband had a neurodevelopmental condition. We excluded trios recruited from Scottish centres and kept unrelated trios. We then split trios into those with both parents unaffected and those with one or both parents affected. These were then categorized as genetically diagnosed or undiagnosed. We applied similar filtering in GEL trios. See Supplementary Methods for more information.
GWAS of neurodevelopmental conditions
We used PLINK v.1.9 to conduct a GWAS comparing individuals with neurodevelopmental conditions (Nâ=â3,618) to controls (Nâ=â13,667) in GEL, controlling for 20 genetic principal components, age and sex. Before running the GWAS, we removed variants with MAFâ<â1%, missingness > 2% or HardyâWeinberg equilibrium Pâ<â1âÃâ10â5, and performed a differential missingness test between the patients with neurodevelopmental conditions and controls and removed variants with Pâ<â1âÃâ10â5. We repeated the GWAS comparing DDD patients with neurodevelopmental conditions on the CoreExome array (Nâ=â6,397) to UKHLS controls (Nâ=â9,270) using PLINK v.1.9, after excluding DDD patients recruited from Scottish centres.
We used METAL80 to conduct an inverse-variance-weighted GWAS meta-analysis between the DDD-UKHLS and GEL GWASs. We removed palindromic SNPs with MAFâ>â0.4 as the strand could not be easily inferred using MAF. We also excluded SNPs with discordant allele frequency (difference > 0.05) between the two cohorts. This left 5,451,801 overlapping SNPs in the meta-analysis.
Heritability
We used several methods to estimate the SNP heritability (the fraction of phenotypic variance explained by genome-wide common variants) on the liability scale assuming a cumulative population prevalence of 1% for rare neurodevelopmental conditions2. First, we applied two methods to estimate SNP heritability using individual-level data in DDD and GEL separately. We performed GREML-LDMS81 stratified by linkage disequilibrium (LD; two bins of equal size) and MAF (three bins: 1â5%, 5â10%, >10%). We also ran phenotype correlationâgenotype correlation (PCGC) regression82, using the LDAK-Thin Model to compute the kinship matrix using the direct method. We corrected for sex, and ten genetic principal components as covariates in both methods. We then meta-analysed the SNP heritability estimates from DDD and GEL using an inverse-variance-weighted method. We also used linkage disequilibrium score regression (LDSC)83 to estimate SNP heritability using summary statistics from the GWAS of neurodevelopmental conditions in DDD, in GEL, and a meta-analysis of the two cohorts. We used roughly 1âmillion common SNPs from HapMap3 with precomputed LD scores. We used the effective sample size (4/(1/Ncasesâ+â1/Ncontrols)) or the sum of two effective sample sizes for the meta-analysis and a sample prevalence of 50% in LDSC, as recommended previously84. We presented the GREML-LDMS estimate in the results, because the estimates were similar to PCGC, and LDSC estimates are known to be under-estimated, especially at low sample size. All estimates are reported in Supplementary Table 3.
Genetic correlations
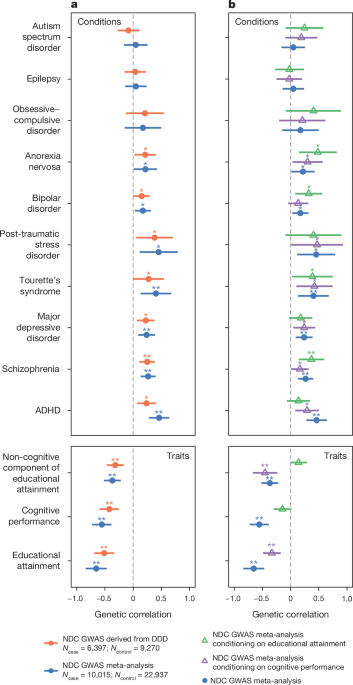
We used LDSC to estimate genetic correlations between the DDD GWAS or the meta-analysed GWAS for neurodevelopmental conditions and various brain-related traits and conditions listed in Supplementary Table 17. We did not use the GEL GWAS to calculate genetic correlations as the SNP heritability was not significantly different from zero according to LDSC.
To estimate the genetic correlations between neurodevelopmental conditions and various brain-related traits or conditions independent of cognitive performance or educational attainment signals, we used genomic structural equation modelling (GenomicSEM)35,85. We estimated the genetic correlation between the target trait and a latent variable representing the non-cognitive component of neurodevelopmental conditions, which was genetic influences on neurodevelopmental conditions that were not captured in the GWAS for cognitive performance31. We applied the GenomicSEM model without SNP effects. We also estimated genetic correlation with the ânon-educational attainmentâ latent variable, which represented genetic influences on neurodevelopmental conditions that were not accounted for by the educational attainment latent variable. We also used GenomicSEM to estimate the percentage of the genetic correlation between neurodevelopmental conditions and the target trait that was explained by latent variables, namely the cognitive and non-cognitive components of neurodevelopmental conditions when conditioning on the cognitive performance GWAS, or EA and non-EA components of neurodevelopmental conditions when conditioning on the educational attainment GWAS (Supplementary Fig. 1 and Extended Data Fig. 9bc). The GenomicSEM model and formulae used to estimate these percentages can be found in Supplementary Fig. 17 and Supplementary Methods.
Calculating polygenic scores
For calculating polygenic scores, we used the set of SNPs that were well imputed in all array cohorts (Minimac4 R2â>â0.8), passed QC in GEL aggV2 samples, and had MAFâ>â1% in all cohorts. We used LDPred86 to estimate weights for calculating polygenic scores and an LD reference panel composed of HapMap3 (ref. 87) common variants based on 5,000 unrelated individuals of genetically inferred White British ancestry from the UK Biobank88 (Supplementary Methods). GWAS summary statistics for years of schooling (a measure for EA)31, the non-cognitive component of educational attainment (NonCogEA)35, cognitive performance (CP)31, schizophrenia (SCZ)32 and neurodevelopmental conditions2 were matched with the list of overlapping SNPs (Supplementary Table 17). PGSNDC,DDD was evaluated in the DDD OmniChip samples and the GEL samples that were not in the DDD GWAS. To make polygenic scores comparable across cohorts (DDD, GEL, UKHLS, MCS and ALSPAC), we performed a joint PCA across all cohorts and adjusted the raw scores for 20 principal components. For most analyses and unless noted otherwise, residuals were scaled so that the combined set of unrelated control samples from GEL and UKHLS (or GEL controls only for PGSNDC,DDD) had mean of 0 and s.d. of 1, and the resultant scores were used for all analyses unless otherwise indicated. In Fig. 3b and Extended Data Fig. 5, we instead show principal component-adjusted polygenic scores that were standardized using weighted MCS average polygenic scores that should represent an unbiased estimate representative of the background population (Supplementary Methods). We also constructed composite polygenic scores combining individual polygenic scores (Supplementary Methods).
Analyses of polygenic scores
Evaluating variance explained by polygenic score
We evaluated how much variance in risk of neurodevelopmental conditions was explained by the polygenic score on the liability scale82,89,90. We compared 6,397 probands with neurodevelopmental conditions from DDD to 9,270 controls from UKHLS, and 3,618 probands with neurodevelopmental conditions from GEL to 13,667 GEL controls defined as described above. We assumed the population prevalence of neurodevelopmental conditions to be 1% (ref. 2).
Comparing polygenic scores between different subsets
We used two-sided t-tests to compare polygenic scores between different groups of probands, parents and controls seen in Figs. 2a and 3b, Extended Data Figs. 5 and 6 and Supplementary Tables 5â7. We report the mean difference in principal component-corrected polygenic scores between groups. Groups who were compared with each other include:
-
Combined set of controls from GEL and UKHLS
-
Control individuals from UK birth cohorts, ALSPAC and MCS
-
Undiagnosed neurodevelopmental condition (NDC) probands regardless of trio status
-
Diagnosed NDC probands regardless of trio status
-
Undiagnosed NDC probands for whom both parents are unaffected
-
Unaffected parents of undiagnosed NDC probands
-
Undiagnosed NDC probands with one or both parents affected
-
Affected parents of undiagnosed NDC probands
-
Diagnosed NDC probands for whom both parents are unaffected
-
Unaffected parents of diagnosed NDC probands
-
NDC probands with de novo diagnoses for whom both parents are unaffected
-
Unaffected parents of NDC probands with de novo diagnoses
-
Diagnosed NDC probands with one or both parents affected
-
Affected parents of diagnosed NDC probands.
Note that âundiagnosedâ and âdiagnosedâ here indicate whether the patient has a monogenic diagnosis. The sample size of each subset is listed in Supplementary Table 1. We excluded controls from UKHLS as well as DDD CoreExome and GSA probands when testing the DDD-derived polygenic score for neurodevelopmental conditions (as these had been included in the original GWAS, whereas the individuals genotyped on the OmniChip had not). All the t-tests involving probands with a neurodevelopmental condition or their parents were performed in samples from DDD and GEL combined.
We also compared female probands versus male probands without a monogenic diagnosis regardless of trio status (2,427 and 1,574 male probands from DDD and GEL, and 1,426 and 918 female probands from DDD and GEL), and unaffected mothers versus unaffected fathers (1,523 trios from DDD and 1,343 trios from GEL) using two-sided t-tests (Extended Data Fig. 8ab).
Polygenic score and diagnostic status
We compared average polygenic scores in probands with a neurodevelopmental condition with and without a monogenic diagnosis using two-sided t-tests, combining probands from DDD and GEL regardless of whether they were in a trio or not. We compared subgroups from families affected by neurodevelopmental conditions to the combined control set from UKHLS and GEL, as well as to unrelated children from the MCS cohort who were reweighted using available sociodemographic data to make them more representative of the general UK population (Supplementary Note 4).
Within DDD (Nâ=â7,549 without excluding Scottish samples or samples who were related to GEL participants), we tested whether the probandâs PGSEA was associated with factors affecting getting a diagnosis in linear regression models:
$${\rm{PGS}}\sim {\rm{factor}}$$
Note that we use the tilde symbol to indicate that the variable before the tilde was regressed on the variable(s) after the tilde. We investigated the following binary factors: trio status (Nâ=â5,507 with both parents exome sequenced but not necessarily genotyped), proband sex (Nâ=â4,421 male probands), whether the proband had any affected first-degree relatives (Nâ=â1,623), whether the proband was born preterm (Nâ=â1,098 with gestation <37âweeks), whether the mother had diabetes (Nâ=â242) and whether the proband had severe intellectual disability or developmental delay (ID/DD; Nâ=â941) versus mild or moderate ID/DD (Nâ=â1,887). We compared probands with the above-mentioned characteristics to all other probands, except when comparing probands with severe versus mild or moderate ID/DD for which we excluded probands without ID/DD or with ID/DD of unknown severity. We also investigated a continuous factor, the degree of consanguinity, quantified by the fraction of the genome in runs of homozygosity (FROH) divided by 0.0625, which is the expected fraction given a first-cousin marriage.
We also tested whether the motherâs or fatherâs PGSEA was associated with the above factors, in a total of 2,497 samples; we did not test for association with trio status as parental genotype data were only available for full trios anyway.
To assess how the association between the above-mentioned factors and diagnostic status changed after correcting for probandâs PGSEA, as well as how the association between probandâs PGSEA and diagnostic status changed after controlling for these factors, we fitted the following logistic regression models:
$${\rm{Diagnostic\; status}}\sim {\rm{factor}}$$
$${\rm{Diagnostic\; status}}\sim {\rm{PGS}}$$
$${\rm{Diagnostic\; status}}\sim {\rm{PGS}}+{\rm{factor}}$$
We also fitted a joint model to assess the effect of PGSEA on diagnostic status controlling for both trio status and prematurity, which showed significant associations with both PGSEA and diagnostic status. We excluded from this joint model factors that were not associated with PGSEA or diagnostic status within the DDD samples with European ancestry (sex, maternal diabetes and FROH), and factors that are likely to be the consequence of having or not having a monogenic diagnosis, rather than a cause of getting a diagnosis (severity of ID/DD and having affected family members).
See the Supplementary Methods for a description of estimation of the odds ratio of diagnosis for different configurations of affected relatives shown in Extended Data Fig. 7a.
Evaluating over-transmission of polygenic scores
We conducted polygenic transmission disequilibrium tests (pTDTs) in undiagnosed and diagnosed probands from DDD (Nâ=â1,523 undiagnosed, 443 diagnosed) and GEL (Nâ=â1,343 undiagnosed, 507 diagnosed) combined. We also conducted pTDTs in these trios excluding autistic probands.
The pTDT is a two-sided one-sample t-test of the probandsâ polygenic score deviation from expectation, which is their parentsâ mean polygenic score. The pTDT deviation is defined as:
$${\rm{pTDT}}\;{\rm{deviation}}={{\rm{PGS}}}_{{\rm{child}}}-({{\rm{PGS}}}_{{\rm{mother}}}+{{\rm{PGS}}}_{{\rm{father}}})/2$$
To evaluate whether the pTDT deviation is significantly different from 0, the pTDT test statistic (tpTDT) is defined as:
$${t}_{{\rm{pTDT}}}=\frac{{\rm{mean}}({\rm{pTDT}}\;{\rm{deviation}})}{\frac{{\rm{s.d.}}({\rm{pTDT}}\;{\rm{deviation}})}{\sqrt{n}}}$$
Association with non-transmitted alleles
Alleles in parents that are not transmitted to the child can still influence the childâs phenotype by affecting the parentsâ behaviour. This phenomenon is called genetic nurture or indirect genetic effects4,26,30. Alleles that are transmitted to the child can influence the childâs phenotype both directly (direct genetic effects) and indirectly through other relatives who carry the same alleles (indirect genetic effects) and whose behaviour is influenced by those alleles. Kong et al. proposed to estimate the direct genetic effect as δâ=âθTâââθNT, where θT indicates the effect of parental transmitted alleles and θNT indicates the effect of parental non-transmitted alleles, which capture both the indirect genetic effects and potential confounding factors4,91. We can estimate θT and θNT of a given polygenic score in the following regression model:
$${{\rm{c}}{\rm{h}}{\rm{i}}{\rm{l}}{\rm{d}}}^{{\prime} }\,{\rm{s}}\,{\rm{p}}{\rm{h}}{\rm{e}}{\rm{n}}{\rm{o}}{\rm{t}}{\rm{y}}{\rm{p}}{\rm{e}}\sim {\hat{\theta }}_{{\rm{T}}}\times {{\rm{P}}{\rm{G}}{\rm{S}}}_{{\rm{T}}}+{\hat{\theta }}_{{\rm{N}}{\rm{T}}}\times {{\rm{P}}{\rm{G}}{\rm{S}}}_{{\rm{N}}{\rm{T}}}$$
where PGST is a polygenic score calculated using transmitted alleles (which is the childâs polygenic score), and PGSNT is a polygenic score calculated using parental non-transmitted alleles, which is equivalent to the difference between the sum of parentsâ polygenic scores and the childâs polygenic score. This model can also be rewritten as:
$$\begin{array}{l}{{\rm{child}}}^{{\prime} }\,{\rm{s}}\,{\rm{phenotype}}\sim ({\widehat{\theta }}_{{\rm{T}}}-{\widehat{\theta }}_{{\rm{NT}}})\,\times \,{{\rm{PGS}}}_{{\rm{child}}}\\ \,\,\,\,\,\,\,\,\,+\,{\widehat{\theta }}_{{\rm{NT}}}\times ({{\rm{PGS}}}_{{\rm{mother}}}+{{\rm{PGS}}}_{{\rm{father}}})\end{array}$$
Therefore, in a regression model in which the childâs polygenic score and parentsâ polygenic scores are both fitted, the coefficient on the childâs polygenic score captures the direct genetic effect, and the coefficient on parentsâ polygenic scores captures the association between non-transmitted alleles and the childâs phenotype. The latter may reflect true indirect genetic effects as well as confounding effects such as uncorrected population stratification and parental assortment29. Thus, we refer to the coefficients on parentsâ polygenic scores in this model as ânon-transmitted coefficientsâ rather than simply âindirect genetic effectsâ, following Young et al.24, as they are mathematically equivalent to the coefficients on the polygenic score constructed from the non-transmitted alleles in a joint regression with the probandâs polygenic score.
We evaluated direct genetic effects (\(\hat{\delta }\)) and effects of maternal and paternal non-transmitted common alleles (\({\widehat{\theta }}_{m,{\rm{NT}}}\) and \({\widehat{\theta }}_{f,{\rm{NT}}}\)) on case status in the following trio model using logistic regression on polygenic scores:
$${1}_{{\rm{N}}{\rm{D}}{\rm{C}}{\rm{s}}{\rm{t}}{\rm{a}}{\rm{t}}{\rm{u}}{\rm{s}}}\sim \hat{\delta }\times {{\rm{P}}{\rm{G}}{\rm{S}}}_{{\rm{c}}{\rm{h}}{\rm{i}}{\rm{l}}{\rm{d}}}+{\hat{\theta }}_{m,{\rm{N}}{\rm{T}}}\times {{\rm{P}}{\rm{G}}{\rm{S}}}_{{\rm{m}}{\rm{o}}{\rm{t}}{\rm{h}}{\rm{e}}{\rm{r}}}+{\hat{\theta }}_{f,{\rm{N}}{\rm{T}}}\times {{\rm{P}}{\rm{G}}{\rm{S}}}_{{\rm{f}}{\rm{a}}{\rm{t}}{\rm{h}}{\rm{e}}{\rm{r}}}$$
where 1NDC status is an indicator variable for whether the individual is a case with a neurodevelopmental condition (1) or control (0). We also ran the regression without correcting for parentsâ polygenic scores (proband-only model) in the same samples for comparison:
$${1}_{{\rm{N}}{\rm{D}}{\rm{C}}{\rm{s}}{\rm{t}}{\rm{a}}{\rm{t}}{\rm{u}}{\rm{s}}}\sim {\hat{\theta }}_{{\rm{T}}}\times {{\rm{P}}{\rm{G}}{\rm{S}}}_{{\rm{c}}{\rm{h}}{\rm{i}}{\rm{l}}{\rm{d}}}$$
Probands with a neurodevelopmental condition were from DDD and GEL trios where the proband was undiagnosed and both parents were unaffected (Nâ=â2,866 trios). Control samples were trios from the two birth cohorts (ALSPAC and MCS, Nâ=â1,434 and Nâ=â2,498, respectively) as well as trios from GEL where the proband did not have DDD-like developmental disorders or neurodevelopmental conditions (Nâ=â872).
We verified that the polygenic scores in the trio model did not show excessive collinearity (Supplementary Methods).
We performed various sensitivity analyses in the following subsets (Supplementary Fig. 4): patients versus controls from GEL trios only, and patients from GEL and DDD versus each of the three control cohorts separately (GEL, MCS or ALSPAC). We also conducted the analysis while controlling for the rare variant burden score (RVBS) in GEL trios (Extended Data Fig. 10b; section below on âAnalyses of polygenic scores and rare coding variantsâ).
$$\begin{array}{c}{1}_{{\rm{N}}{\rm{D}}{\rm{C}}{\rm{s}}{\rm{t}}{\rm{a}}{\rm{t}}{\rm{u}}{\rm{s}}}\sim {{\rm{P}}{\rm{G}}{\rm{S}}}_{{\rm{c}}{\rm{h}}{\rm{i}}{\rm{l}}{\rm{d}}}+{{\rm{R}}{\rm{V}}{\rm{B}}{\rm{S}}}_{{\rm{c}}{\rm{h}}{\rm{i}}{\rm{l}}{\rm{d}}}+{{\rm{P}}{\rm{G}}{\rm{S}}}_{{\rm{m}}{\rm{o}}{\rm{t}}{\rm{h}}{\rm{e}}{\rm{r}}}\\ \,\,\,\,\,+\,{{\rm{R}}{\rm{V}}{\rm{B}}{\rm{S}}}_{{\rm{m}}{\rm{o}}{\rm{t}}{\rm{h}}{\rm{e}}{\rm{r}}}+\,{{\rm{P}}{\rm{G}}{\rm{S}}}_{{\rm{f}}{\rm{a}}{\rm{t}}{\rm{h}}{\rm{e}}{\rm{r}}}+{{\rm{R}}{\rm{V}}{\rm{B}}{\rm{S}}}_{{\rm{f}}{\rm{a}}{\rm{t}}{\rm{h}}{\rm{e}}{\rm{r}}}\end{array}$$
We restricted this latter analysis to GEL trios to minimize artefactual differences in rare variant calling and QC between cases and controls, which could otherwise create spurious associations.
See the Supplementary Methods for a description of how we modified the running of this trio model to investigate the hypothesis that the effects of non-transmitted alleles associated with educational attainment and cognition might be mediated by prematurity.
Analyses of polygenic scores and rare coding variants
Sequence data from DDD, GEL and MCS were annotated with the Ensembl Variant Effect Predictor (VEP)92. We kept the âworst consequenceâ annotation across transcripts. From parents and probands, we extracted autosomal heterozygous PTVs (transcript ablation, frameshift, splice acceptor, splice donor and stop gained) annotated as high-confidence by LOFTEE93 (HC PTVs), as well as variants in the following classes that we grouped as âmissenseâ: missense, stop lost, start lost, inframe insertion, inframe deletion and loss-of-function variants annotated as low-confidence by LOFTEE93. We retained rare variants with MAFâ<â1âÃâ10â5 in each gnomAD super-population and MAFâ<â1âÃâ10â4 in the respective cohorts.
We considered four (non-mutually exclusive) groups of damaging rare variants:
-
1.
HC PTVs in constrained genes (pLIâ>â0.9)94
-
2.
HC PTVs and missense variants (MPCââ¥â2)95 in constrained genes (pLIâ>â0.9)
-
3.
HC PTVs in monoallelic DDG2P genes with a loss-of-function mechanism (that is, âabsent gene productâ)
-
4.
HC PTVs and missense variants (MPCââ¥â2) in monoallelic DDG2P genes with a loss-of-function mechanism.
We retained probands and parents who were heterozygous for these variants. We required the variants in the children to have been inherited from a parent.
To investigate whether parental assortment leads to correlated rare and common variant burden, we calculated rare variant burden scores as the number of rare variants in the classes described above, then calculated the Pearsonâs correlation coefficients between rare variant burden scores and polygenic scores using the âcorâ function in R. We used trios in which both parents were unaffected in this analysis. Rare variant burden scores were corrected for 20 genetic principal components using linear regression models. We then calculated the correlation coefficients between the principal component-adjusted rare variant burden scores in parents and the principal component-adjusted polygenic scores in their partners. We also assessed the correlation within the same person among either children or parents. We repeated the analysis in subsets of trios in which the proband was undiagnosed as well as in trios in which the proband had a monogenic de novo diagnosis (Supplementary Fig. 6). The main analysis in Fig. 5 and the sensitivity analysis in Extended Data Fig. 10b is based on group 2 above, whereas Supplementary Figs. 6â8 show the results for all four groups of variants. To investigate whether the results were affected by uncorrected population structure, we also calculated rare variant burden scores using rare synonymous variants in either monoallelic DDG2P genes with a loss-of-function mechanism or constrained genes, and assessed their correlation with polygenic score.
To assess whether polygenic scores modify penetrance of rare inherited variants, we conducted one-sided paired t-tests comparing the polygenic score between unaffected parents transmitting a damaging variant to their affected offspring who inherited the variant (Supplementary Fig. 8). We hypothesized that the unaffected parents would have a more protective polygenic background than their affected offspring (indicated by higher PGSEA, PGSCP, PGSNonCogEA and lower PGSSCZ, PGSNDC,DDD). If more than one parent transmitted a variant to a proband, one parentâchild pair was chosen at random from the trio. We used trios in which the proband was undiagnosed and both parents were unaffected in this analysis.
Construction and use of weights for MCS
We were concerned that control cohorts might not be random samples of the population with respect to educational attainment, and that this might bias our effect sizes for the difference in polygenic scores between cases and controls (Supplementary Note 4). We decided to use MCS, for which extensive sociodemographic data are available, to calculate a mean polygenic score that would be representative of the general population, using inverse-probability weighting. MCS deliberately oversampled minority ethnic and disadvantaged individuals in the United Kingdom96 (sampling bias), and they provide sampling weights to account for this. Furthermore, missingness in each wave of data collection, including the collection of DNA for genotyping, was non-random (non-response bias). To correct for non-response bias, we produced non-response weights per individual using the inverse of the probability of being genotyped estimated from a logistic regression, considering covariates collected at the first study sweep, as previously described96,97 (Supplementary Methods). We fitted the model to predict who was in the sample of unrelated children of GBR ancestry with genotype data (Nâ=â5,884 of 6,036 children who had complete data for these covariates), and separately to predict who was in the subset of these that also had genotype data on both parents (Nâ=â2,445 of 2,498 trio children who had no missingness). To produce weights that account for both sampling bias and non-response bias, we multiplied the non-response weight from regression models by the sampling weights provided by MCS. These weights were then used to calculate adjusted polygenic scores shown in Fig. 3b and Extended Data Figs. 5 and 6c and adjusted correlation between polygenic score and rare variant burden score shown in Supplementary Fig. 7.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.