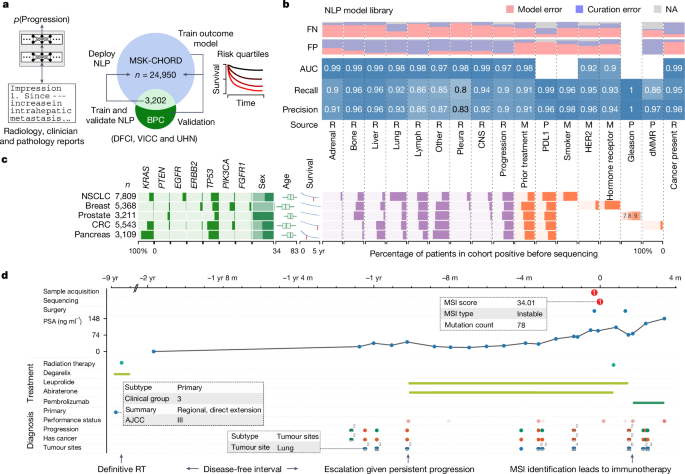
Patients
This study primarily analysed data for patients with tumour genomic sequencing and completed tumour registry entries from two partially overlapping sources: patients with MSK-IMPACT sequencing (forming the basis of MSK-CHORD) and the Project GENIE BPC cohort of the American Association for Cancer Research, which includes patients with tumour genomic profiling and clinical annotation from four institutions including MSK. Details regarding the BPC have been published previously7. Here we included patients in the BPC with single-primary NSCLC, breast, colorectal, prostate or pancreatic cancer. The MSK-CHORD cohort comprises patients at MSK, an academic cancer hospital with tumour genomic sequencing using MSK-IMPACT, a Food and Drug Administration-authorized tumour genomic profiling assay, which uses matched white blood cell sequencing to filter clonal haematopoiesis and germline variants. All MSK patients were enrolled as part of a prospective sequencing protocol (NCT01775072) or analysed as part of institutional review board (IRB)-approved retrospective research protocols (MSK IRB protocols 16â1463 and 19â368). The study was independently approved by the IRBs of MSK and DFCI. Patients provided written, informed consent and were enrolled in a continuous, non-random fashion. Data here are from a 9 September 2023 snapshot.
Genomic annotation
For all analyses involving tumour genomic alterations aggregated at the gene level, a Food and Drug Administration-recognized molecular knowledge database (OncoKB34) was used to annotate all mutations, copy number changes and structural variations as oncogenic or not; any such oncogenic alteration led to a gene being labelled positive for the purposes of analysis. For OS modelling in which non-MSK BPC patients were used as an external validation cohort, only genes present in all sequencing panels across the BPC were used as variables. For other genomic analyses, the 341 genes included in the first MSK-IMPACT sequencing panel20 were used as variables. The presence or absence of genomic gains and losses of each chromosome arm were identified from MSK-IMPACT data. Genomic coordinates for the chromosome arms in the GRCh37 (also known as hg19) human genome assembly were considered gained or lost if most of the arm (>50%) is made up of segments with an absolute value log ratio of â¥0.2 (ref.â44).
NLP models
Radiology reports
Data preprocessing. Radiology reports for computed tomography (CT), positron emission tomography and magnetic resonance imaging examinations of chest, abdomen, pelvis, head and/or extremities were queried for all patients within the MSK-CHORD cohort. Report sections were segmented using regular expression to separate the âimpressionâ section from the full report, for cases in which it was available. The pieces of impression text corresponding to the manually curated MSK-BPC labels for presence of cancer, tumour sites and cancer progression were merged to create a direct mapping of label and text that the labels were derived from.
Radiographic progression. We fine-tuned a RoBERTa transformer model45 on impression sections extracted from radiology reports paired with binarized human-curated progression labels. Labels were binarized by calling the two GENIE BPC label classes âProgressing/Worsening/Enlargingâ and âMixedâ as positive, and calling other classes (âImproving/Respondingâ, âStable/No changeâ and âNot stated/Indeterminateâ) as negative.
Binarized supervision labels were provided at the document level (that is, the model was trained to predict a single binary variable for a given impression section). We used the PyTorch46 implementation of RoBERTa and pretrained model weights from the HuggingFace library and model hub47. Text was tokenized with the default RoBERTa tokenizer and report-level predictions are pooled using the default method of conditioning on the first [CLS] pseudo-token prepended to the sequence comprising the impression section. We used a batch size of 128, fine-tuning with the AdamW optimizer48 using a learning rate of 2âÃâ10â6, and fine-tuning for 20 epochs with a linearly decaying learning-rate scheduler with a 2-epoch warm-up period. Hyperparameter values were selected through a random search using a holdout validation set of 20% of reports across learning rate values {1âÃâ10â6, 2âÃâ10â6, 5âÃâ10â6, 1âÃâ10â6}, batch size values {8, 16, 32, 64, 128, 256} and num-epochs {5, 10, 20, 50}. Extrinsic results (that is, main results incorporating model predictions) were presented on models trained on the full MSK-BPC cohort.
Tumour sites. We fine-tuned a ClinicalBERT model49, which is itself a BioBERT model50 fine-tuned on reports from the MIMIC-III v1.4 database51. We extracted impression sections from radiology reports and paired them with report-level supervision from the GENIE BPC dataset. Labels were transformed into ten binary variables corresponding to a closed inventory of nine common disease sites (adrenal gland, bone, CNS or brain, intra-abdominal, liver, lung, lymph nodes, pleura and reproductive organs), along with one âotherâ variable, describing whether the report is labelled as indicating tumour presence in that organ site.
The model was trained in a multi-labelling setup: pooled transformer output was input to a single-layer fully connected feed-forward network of width d with a tanh nonlinearity, whose output is linearly transformed to a ten-dimensional vector giving ten logits, from which binary cross-entropy losses were computed against the gold-standard labels and mean-pooled. In other words, the network computes
$$f(x)={\sigma }(V(\tanh (W{\varphi }(x))))$$
in which x is the tokenized document, Ï(x) is the pooled transformer output vector, W is a learned affine transformation outputting a d-dimensional vector, tanh is applied element-wise, V is a learned affine transformation mapping d-dimensional vectors to ten-dimensional vectors, and Ï is a plain element-wise sigmoid function; f(x) is a ten-dimensional vector of values between 0 and 1. Note that the different per-site predictions are non-mutually-exclusive and are conditionally independent given the post-pool d-dimensional hidden state.
The ClinicalBERT model was implemented in PyTorch46; we used the model and pretrained model weights in the HuggingFace library and model hub47. We pooled transformer model output using the default method of conditioning on the first [CLS] pseudo-token prepended to the sequence comprising the impression section. We trained using AdamW48 trained using a batch size of 8, a learning rate of 2âÃâ10â6, a dropout probability of 0.2 (applied to the post-pool single-hidden-layer feed-forward network) and a pre-logit hidden dimension of 1,024, training for 15 epochs with a warm-up period of 1.5 epochs. Extrinsic results (that is, main results incorporating model predictions) were presented on models trained on the full MSK-BPC cohort.
Cancer presence. We fine-tuned a BERT52 base model (uncased)53 on impression sections extracted from radiology reports paired with binarized human-curated cancer evidence labels. Labels were binarized by calling the MSK-BPC label class 1 as âyesâ for presence of cancer and calling label class 0 as ânoâ for absence of cancer. Binarized supervision labels were provided at the document level (that is, the model was trained to predict a single binary variable for a given impression section). BERT models were trained as described for tumour sites. Text was tokenized with the default HuggingFace AutoTokenizer for BERT, and report-level predictions were pooled using the default method of conditioning on the first [CLS] pseudo-token prepended to the sequence comprising the impression section. We used a batch size of 32 and fine-tuned for a maximum of 10 epochs. We trained the models using the AdamW optimizer48 using a learning rate of 1âÃâ10â5, epsilon of 1âÃâ10â8, weight decay of 1âÃâ10â4 and no warm-up period. During training, model weights were optimized to minimize cross-entropy loss.
Clinician notes
Data preprocessing. Clinician notes for patients were queried and filtered by initial consult (IC) and follow-up notes created by medical oncologists, radiation oncologists, surgery, inpatient services and others. Notes in the institutional database are segmented into subsections including family history, present illness, comorbidities and so on. Further filtering or combining of note subsections was dependent on the application. For inferring prior outside medications, IC notes were filtered and included sections relevant to external treatments, such as past medical history, history of present illness and chief complaint, while excluding sections mentioning future treatment plans. Patients with no IC notes in the allowable note categories were excluded from the training and validation set. We excluded patients with IC notes dated more than 90 days after their initial visit date. We selected one note per patient to analyse. If a patient had multiple notes, the IC note with the earliest creation time was used. Preprocessing for inference of HER2 and hormone receptor consisted of filtering notes created by the breast medicine division, for which entire IC and follow-up notes were used as the input to the model.
Prior external treatment. The other transformer-based models presented above operate on impression sections that are generally very short and therefore do not see appreciably degraded marginal performance from truncating documents to the maximum model input sequence size of 512 subtokens. This relatively short input limit is necessary for the full-self-attention parameterizations used by these models, which require memory scaling quadratically in input sequence length. However, full IC reports are appreciably longer than impression sections, and any mention of prior anti-neoplastic treatments occurs within a much longer textual context. We therefore use a transformer model engineered to have subquadratic memory requirements; in particular, we fine-tune a Clinical-Longformer model54, which is itself a Longformer model55 fine-tuned on the MIMIC-III v1.4 database51. This model has a maximum input sequence length of 4,096 subtokens.
The Clinical-Longformer model is implemented in PyTorch46; we use the model and pretrained model weights in the HuggingFace library and model hub47. We pool transformer model output using the default method of conditioning on the first [CLS] pseudo-token prepended to the sequence comprising the impression section. We train AdamW48 using a batch size of 64 and a learning rate of 1âÃâ10â6, training for 20 epochs with a warm-up period of 2 epochs. We upsample minority-class examples uniformly with replacement to achieve class balance during training. Extrinsic results (that is, main results incorporating model predictions) are presented on models trained on the MSK-BPC cohort.
HER2 and hormone receptor. As HER2 and hormone receptor can be heterogeneous across pathology samples, we sought to create a classifier based on clinician notes to identify the overall receptor subtypes for a patientâs cancer used to inform treatment. For training, we used clinician notes from a cohort of 6,053 patients with single-primary breast cancer with manually annotated HER2 and hormone receptor subtypes to train separate HER2 and hormone receptor binary classifiers. We performed training and testing within this cohort with a 90/10 split. Specifically, the clinician note chronologically closest to sequencing was used as features and the expert-annotated subtypes as targets. For final validation, we used a held-out set of 1,489 patients from a previously published breast cancer dataset22. As with the prior treatment model, we used Clinical-Longformer models for both HER2 and hormone receptor classifiers using a 2,000-subtoken input, padded as necessary. We used the AdamW optimizer with a batch size of 64 and a learning rate of 1âÃâ10â6, training for 30 epochs without a warm-up period.
Smoking status. Smoking status (former or current versus never) was obtained from dedicated smoking or social history sections through regular expression extraction applied to the first available clinician assessment for a given patient. The algorithm was created on the basis of a previously published cohort of 247 patients with NSCLC and previously annotated smoking status14, withholding data from patients also present in the MSK-BPC NSCLC cohort. The model was validated on the basis of the MSK BPC NSCLC cohort.
Pathology reports
PDL1. PDL1 status (positive defined as 1% or higher versus negative) was obtained through regular expression extraction applied to the first available clinician assessment for a given patient. The algorithm was created on the basis of a previously published cohort of 247 patients with NSCLC and previously annotated smoking status14, withholding data from patients also present in the MSK BPC NSCLC cohort. The model was validated on the basis of the MSK BPC NSCLC cohort.
Gleason score. Gleason score (6â10) was obtained through regular expression extraction applied to pathology reports from either prostatic biopsies or resections. The algorithm was created on the basis of iterative fine-tuning on a previously published cohort of 451 patients with prostate cancer and previously annotated Gleason score56, withholding data from patients also present in the MSK BPC Prostate cohort. The model was validated on the basis of the MSK BPC Prostate cohort.
MMR. Mismatch status (proficient versus deficient) was obtained through regular expression extraction applied to histopathology reports. The algorithm was created on the basis of a previously published cohort of 224 patients with CRC and previously annotated MMR status57, withholding data from patients also present in the MSK CRC cohort. The model was validated on the basis of the MSK BPC CRC cohort.
Billing code annotation metrics. We sought to assess the accuracy of structured data elements (that is, billing codes12) to recover tumour site information and to compare this accuracy with that of our NLP algorithms. As the timing of billing codes is not necessarily tied to particular radiology reports, we aggregated labels at the patient level, wherein cancer detection in a given tumour site at any point in the patientâs history was considered positive overall for that site. Patient-level billing code labels and, separately, NLP labels (from radiology impressions as above) were compared to gold-standard curated BPC labels, all aggregated at the patient level. The patient-level accuracies for these annotations are provided in Supplementary Table 2.
OS modelling
RSFs58 to predict time to death from time of cohort entry, right-censored at time of last follow-up, were trained using pre-assigned hyperparameters (n treesâ=â1,000, minimum n splitsâ=â10, minimum n samples per leafâ=â15). In exploratory secondary analyses, a random hyperparameter grid search to find âoptimalâ hyperparameters using a 20% holdout for evaluation was conducted (n tree range 200â2,000, minimum n splits range 5â20, minimum n samples per leaf range 5â30, n search iterationsâ=â100, threefold internal cross-validation for hyperparameter selection); a model trained on optimal hyperparameters did not yield better results (c-index âimprovementâ of â0.01 using optimal versus pre-assigned hyperparameters). We included all variables in Supplementary Table 6, grouped according to the schema in that table.
To predict time to death while accounting for left truncation and right censoring, we used the OncoCast package (https://github.com/AxelitoMartin/OncoCast) updated from previous work59,60 with the RF (Random Forest) method. In brief, this method fits an elastic net-regularized Cox proportional hazards model to the data, and then applies a random forest to estimate the Martingale residuals; this correction term is applied when the model is tested on new data. We created an ensemble learning model through cross-validation or by training on the whole MSK-CHORD dataset and validating the model on the non-MSK BPC dataset as for the RSF model. The OncoCast model, configured with 500 trees, 5 terminal nodes and 50 runs, was fitted to the training set. Predictions of risk for the test set were made across all iterations. Model performance was assessed using the concordance probability index at each iteration.
radLongformer
We fine-tuned a Clinical-Longformer54 model to take as input the full text of CT chest, abdomen and pelvis (CAP) reports and predict binarized OS within 6 months, a clinically meaningful endpoint and a time frame in which a single radiology report might meaningfully prognosticate. We split all cohorts into training and test sets at the patient level, reserving 20% of the cohort or all patients with a CT CAP within 3 months of cohort entry for testing, whichever was smaller. In the training set, all CT CAP reports from all patients were annotated according to survival status within 6 months; those with insufficient follow-up were excluded. The Clinical-Longformer was fine-tuned in this dataset using a batch size of 64 and a learning rate of 1âÃâ10â6, training for 20 epochs with a warm-up period of two epochs.
Time to metastasis
The association of genomic alterations with time to metastasis was analysed using Cox proportional hazards models. Death was treated as a censoring event. Patients with metastasis to a given site of interest before the start time (time of sample acquisition; that is, the earliest time a given alteration could be confirmed for a given tumour) were excluded from analysis. Prior treatment (any versus none) and stage (IâIII versus IV) were included as variables in all multivariable analyses. Histologic subtype was included as a variable where indicated.
SETD2 validation cohorts
We utilized two validation cohorts of patients with LUAD and tumour genomic profiling: patients at DFCI; and patients in a commercial real-world dataset. Details of the DFCI cohort have been published previously1,61. In the commercial dataset, formalin-fixed paraffin-embedded samples from patients with NSCLC were submitted to a commercial Clinical Laboratory Improvement Amendments-certified laboratory for molecular profiling (Caris Life Sciences, Phoenix, AZ). Any patient with Caris tumour molecular profiling was eligible for inclusion; patient sources include a variety of community and academic settings, and patients were non-overlapping with those in MSK-CHORD. A total of 29,422 NSCLCs with adenocarcinoma histology were analysed by next-generation sequencing, 592 targeted panel or whole-exome sequencing for genomic features. Before molecular testing, tumour enrichment was achieved by collecting targeted tissue using manual microdissection techniques. For NextSeq-sequenced tumours, a custom-designed SureSelect XT assay was used to enrich 592 whole-gene targets (Agilent Technologies, Santa Clara, CA). For NovaSeq whole-exome-sequenced tumours, a hybrid pull-down panel of baits designed to enrich for more than 700 clinically relevant genes at high coverage and high read depth was used, along with another panel designed to enrich for an additional >20,000 genes at a lower depth. A 500-megabase single-nucleotide polymorphism backbone panel (Agilent Technologies, Santa Clara, CA) was added to assist with gene amplification and deletion measurements and other analyses. All variants were detected with >99% confidence, with an average sequencing depth of coverage of >500 and an analytic sensitivity of 5%. This test has a sensitivity to detect as low as approximately 10% population of cells containing a mutation in all exons from the high-read-depth clinical genes and 99% of all exons in the 20,000 whole-exome regions. Genetic variants identified were interpreted by board-certified molecular geneticists and categorized according to the American College of Medical Genetics and Genomics standards. Real-world OS was obtained from insurance claims data and calculated from time of biopsy to last contact.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.