Mice
Adult C57BL/6J wild-type male mice from Jackson Laboratories were used in all experiments except for inhibitory tagging experiments (Extended Data Figs. 5 and 6). In those experiments, Gad2–cre male mice from Jackson Laboratories (or bred in-house from Jackson Laboratories) were used. Mice ordered from Jackson arrived group-housed in cages of 4 mice per cage and were singly housed for the experiment. Mice underwent behavioural testing at 12â18âweeks of age. For experiments in which mice underwent PSAM virus injections, mice were included in the experiment if there was expression of GFP+ cell bodies in both the dorsal and ventral hippocampus. All experimental procedures were approved by the Icahn School of Medicine at Mount Sinaiâs IACUC.
Viral constructs
For calcium imaging experiments in Figs. 2â6 and Extended Data Figs. 3, 4 and 7â10, AAV1-Syn-GCaMP6f-WPRE-SV40 (titre, 2.8âÃâ1013 genome copies per ml) was purchased from AddGene and was diluted by 4 in sterile 1à PBS (final titre, ~7âÃâ1012 genome copies per ml). The mice had 300ânl of the diluted virus injected into the right hemisphere of the dorsal CA1. For PSAM experiments, AAV5-Syn-PSAM4-GlyR-IRES-eGFP (2.4âÃâ1013 genome copies per ml) was purchased from AddGene. Mice had the virus injected at stock titre bilaterally into the dorsal and ventral hippocampus, 300ânl per injection site. For inhibitory tagging experiments, a virus cocktail of AAV1-Syn-GCaMP6f-WPRE-SV40 (titre, 1.3âÃâ1013 genome copies per ml) and AAV5-hSyn-DIO-hM3Dq-mCherry (titre, 2.6âÃâ1013 genome copies per ml) (both purchased from AddGene) was mixed 1:1 and mice had 300ânl of this mixed virus cocktail injected into the right hemisphere of the dorsal CA1.
Surgery
Mice were anaesthetized with 1 to 2% isoflurane for surgical procedures and placed into a stereotaxic frame (David Kopf Instruments). Eye ointment was applied to prevent desiccation, and the mice were kept on a heated pad to prevent hypothermia. Surgery was performed using aseptic technique. After surgery, carprofen (5âmg per kg) was administered every day for the following 3 days, and ampicillin (20âmg per kg) was administered every day for the next 7âdays. For calcium imaging experiments, dexamethasone (0.2âmg per kg) was also administered for the following 7âdays.
For PSAM experiments (Extended Data Fig. 1lâp), AAV5-Syn-PSAM4-GlyR-IRES-eGFP was injected at stock concentration. Mice had 300ânl of the virus injected bilaterally into the dorsal hippocampus (anteroposterior (AP), â2âmm; mediolateral (ML), ±1.5âmm; dorsoventral (DV), â1.5âmm) and 300ânl injected bilaterally into the ventral hippocampus (AP, â3âmm; ML, ±3.2âmm; DV, â4âmm), for a total of four injections and 1.2âμl injected per mouse, using a glass pipette and the Nanoject injector. The pipette was slowly lowered to the injection site, the virus was injected at 2ânlâsâ1 and then the pipette remained for 5âmin before being removed to allow diffusion of the virus. Mice had their incision sutured after surgery and betadine was applied to the site to prevent infection.
For calcium imaging experiments in Figs. 1â4 and 6, mice underwent two serial procedures spaced 1âmonth apart, as described previously29. During the first surgery, a 1âmm diameter craniotomy was made above the dorsal hippocampus on the right hemisphere (centred at AP, â2âmm; ML, +1.5âmm from bregma). An anchor screw was screwed into the skull on the contralateral hemisphere at approximately AP â1âmm and ML â2.5âmm from bregma. Then, 300ânl of AAV1-Syn-GCaMP6f was injected into dorsal CA1 of the hippocampus on the right hemisphere (AP, â2âmm; ML, +1.5âmm; DV, â1.2âmm). Virus was injected as described in the PSAM experiments above. After the pipette was removed, the mouse remained on the stereotaxic frame for 20âmin to allow complete diffusion of the virus. After 20âmin of diffusion, the cortex below the craniotomy was aspirated with a 27-gauge blunt syringe needle attached to a vacuum pump, while constantly being irrigated with cortex buffer. When the striations of the corpus callosum were visible, the 27-gauge needle was replaced with a 30-gauge needle for finer-tuned aspiration. Once most of corpus callosum was removed, bleeding was controlled using surgical foam (Surgifoam), and then a 1âmm diameter à 4âmm length GRIN lens (GRINTECH) was slowly lowered into the craniotomy. The lens was fixed with cyanoacrylate, and then dental acrylic was applied to cement the implant in place and cover the rest of the exposed skull. The top of the exposed lens was covered with Kwik-Sil (World Precision Instruments) to protect it and the Kwik-Sil was covered with dental cement. Then, 4âweeks later, the mice were again put under anaesthesia to attach the baseplate, visually guided by a Miniscope. The overlying dental cement was drilled off and the Kwik-Sil was removed to reveal the top of the lens. The Miniscope with an attached baseplate was lowered near the implanted lens and the field of view was monitored in real-time on a computer. The Miniscope was rotated until a well-exposed field of view was observed, at which point the baseplate was fixed to the implant with cyanoacrylate and dental cement. The mouse did not receive post-operative drugs after this surgery as it was not invasive. For inhibitory tagging experiments, the surgeries were performed as described above; however, they were separated into three surgeries rather than two: first, the virus injection was done and the mice had the incision sutured after the surgery. The lens implant procedure was done during a separate surgery 1â7 days later. Baseplating was done 1âmonth after viral injection during a third surgery.
For calcium imaging experiments with EEG/EMG implants (Fig. 5 and Extended Data Figs. 9 and 10), mice underwent three serial procedures spaced around 2âweeks apart. During the first surgery, mice had 300ânl of AAV1-Syn-GCaMP6f injected into dorsal CA1 as described above, but the incision was sutured after the surgery. Then, 2âweeks later during a second surgery, mice had their overlying cortex aspirated and a GRIN lens was implanted above the injection site, as above. During this surgery, a wireless telemetry probe (HD-X02, Data Science International) was also implanted with EEG and EMG wires. Two EMG wires were implanted into the left trapezius muscle. One EEG wire was implanted between the skull and dura mater above the dorsal hippocampus on the contralateral hemisphere to the GRIN lens (left hemisphere; AP, â2âmm; ML, â1.5âmm), and a reference EEG wire was implanted between the skull and the dura on the right hemisphere overlying the prefrontal cortex (AP, +1.75âmm; ML, â0.5âmm). Cyanoacrylate and dental cement fixed the GRIN lens, anchor screw and EEG wires in place. The telemetry probes were implanted during the second surgery rather than the first to minimize the time that the mice needed to live with the implant (because the mice sometimes reject the implant after long periods). During the third procedure, the mice were returned to implant the baseplate, as described above.
Behavioural procedures
Before all of the experiments, the mice were handled for 1âmin each day for at least 1âweek. On at least four of those days, the mice were transported to the testing room and handled there. On the rest of the days, the mice were handled in the vivarium. In calcium imaging experiments, mice were handled and habituated for 2 weeks instead of 1, during which they were habituated to having the Miniscope attached and detached from their heads. To become accustomed to the weight of the Miniscope, they were placed in their home cage with the Miniscope attached for 5âmin per day for at least 5 days.
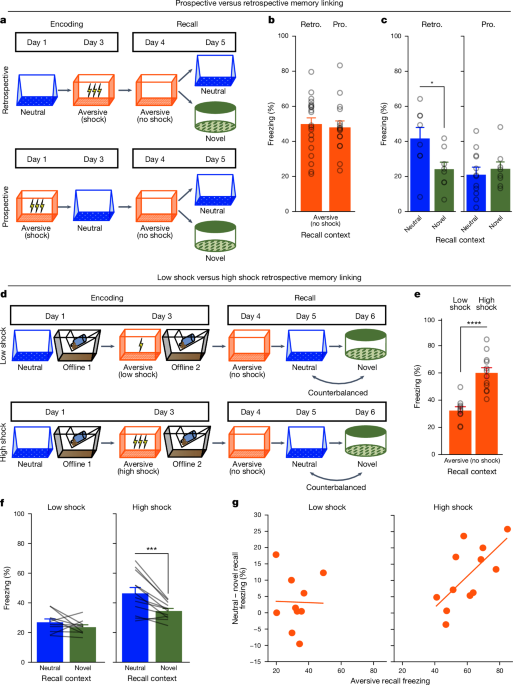
In memory-linking behavioural experiments, mice were exposed to the neutral context for 10âmin to explore. During aversive encoding, after a baseline period of 2âmin, mice received three 2âs foot shocks of either amplitude 0.25âmA (low-shock) or 1.5âmA (high-shock), with an intershock interval of 1âmin. Then, 30âs after the final shock, the mice were removed and returned to the vivarium. On the next 3 days, the mice were tested in the previously experienced aversive and neutral contexts, as well as a completely novel context that they had not been exposed to previously, for 5âmin each. The features of the neutral and novel contexts were counter-balanced and were made up of different olfactory, auditory, lighting and tactile cues. The aversive context was always the same with distinct cues from the neutral and novel contexts. In the PSAM experiment (Extended Data Fig. 1lâp), the mice were tested in either the aversive, neutral or novel context. In the prospective versus retrospective memory-linking experiment (Fig. 1aâc), mice were tested in the aversive context first, and then half of the mice were tested in the neutral context and the other half in the novel context. In the low- versus high-shock experiments (Fig. 1dâg and Extended Data Figs. 1câe and 9b,c), mice were tested in the aversive context first, followed by testing in the neutral and novel context counter-balanced; half of the mice received neutral recall and then novel-context exposure the next day, and the other half received novel-context exposure and then neutral recall. All testing was done in Med Associates chambers. Behavioural data were processed using the Med Associates software for measuring freezing. In experiments in which mice were tethered with a Miniscope, behavioural data were processed using our previously published open-source behavioural tracking pipeline, ezTrack61 v.1.2. In the prospective versus retrospective memory-linking temporal window experiments (Extended Data Fig. 1a,b), the aversive learning experience was distinct: mice explored for 2âmin, then administered one 0.75âmA, 2âs foot shock and removed from the context 30âs after this shock.
In cocaine retrospective memory-linking experiments (Extended Data Fig. 2), mice were placed in the same contexts that were used in the above aversive memory-linking experiments (that is, Med Associates chambers). For cocaineâcontext pairings, mice were injected with cocaine (or saline as a control) and immediately placed in the conditioning context for 10âmin. For encoding of the neutral context, mice were placed in the context for 10âmin. Recall sessions were 5âmin each. Behavioural data were processed using the Med Associates software for measuring locomotion.
Drug injections
For PSAM experiments (Extended Data Fig. 1lâp), uPSEM-817 tartrate was made in a solution of 0.1âmgâmlâ1 in saline and injected intraperitoneally at a dose of 1âmg per kg (10âmlâkgâ1 injection volume). Previous studies have shown that PSAM4-GlyR (PSAM), an inhibitory ionotropic receptor with no endogenous ligand, binds with the injectable PSEM ligand to cause robust hyperpolarization in neurons62. Saline was used as a vehicle. The first injection was done as soon as the mice were brought back to the vivarium after aversive encoding (around 3âmin after the end of aversive encoding). The next three injections were done every 3âh to cover a 12âh timespan of inhibition. For cocaine retrospective memory-linking experiments, mice were injected with 10âmg per kg (10âmlâkgâ1 injection volume) of cocaine dissolved in saline, or injected with saline as a control. For chemogenetic identification of inhibitory neuron experiments (Extended Data Figs. 5 and 6), clozapine N-oxide dihydrochloride (CNO) was made in a solution of 0.3âmgâmlâ1 in saline and injected intraperitoneally at a dose of 3âmg per kg (10âmlâkgâ1 injection volume). In Extended Data Fig. 5, all of the mice were injected with saline on the first day. On the second day, mice were injected with CNO or saline and, on the third day, mice were injected with saline or CNO, whichever solution they did not receive the day before.
Calcium imaging Miniscope recordings
Open-source V4 Miniscopes (https://github.com/Aharoni-Lab/Miniscope-v4) were connected to a coaxial cable, which was connected to a Miniscope data acquisition board (DAQ) 3.3. The DAQ connected to a computer through USB3.0. Data were collected through the Miniscope QT Software v.1.11 (https://github.com/Aharoni-Lab/Miniscope-DAQ-QT-Software) at 30âfps. The Miniscopes were either assembled in-house or purchased from Open Ephys, and DAQ boards were purchased from Open Ephys.
When performing calcium imaging with concurrent behaviour in the Med Associates boxes, mice were brought into the testing room from the vivarium, taken out of their home cage and had the Miniscope attached. They were placed back into their home cage for 1âmin. They were then removed from their home cage and placed into the testing chamber. To record calcium and behaviour, the Med Associates software sent a continuous TTL pulse to record from the Miniscope while the behaviour was concurrently tracked using Med Associates cameras. After the session was complete, the mice were immediately returned to their home cage, then the Miniscope was removed, and the mouse was returned to the vivarium. One mouse was brought to the testing room at a time.
For calcium imaging experiments without simultaneous EEG and EMG recordings, offline calcium imaging recordings were done in the mouseâs home cage for the 1âh after neutral encoding and after aversive encoding. During these recordings, mice were placed back into their home cage and the home cage was placed into a large rectangular and opaque storage bin to occlude distal cues, with a webcam (Logitech C920e or MiniCAM) overlying the home cage to track behaviour during the recording. Using the Miniscope QT Software with two devices connected (Miniscope and webcam), calcium imaging and behaviour were concurrently tracked. After the offline recording was complete, mice were removed from their home cage, the Miniscope was removed, they were returned to their home cage and returned to the vivarium immediately thereafter. The same procedure was undergone for the experiment in Extended Data Fig. 3. For calcium imaging experiments with simultaneous EEG and EMG recordings, mice lived in a custom-made home cage where offline recordings could take place. These home cages (Maze Engineers) were custom designed to accommodate mice wearing a Miniscope chronically for the duration of the experiment (about 2 weeks total). The water spout and food hopper were side-mounted and there was a slit along the top of the home cage so that the Miniscope coaxial cable could freely move. This home cage was placed on top of a receiver that would wirelessly receive EEG, EMG, temperature and locomotion telemetry data continuously throughout the experiment (HD-X02, Data Science International). Mice had a Miniscope attached on the first day and were allowed to wear it for an hour in their home cage to acclimatize to its weight, after which it was removed. On the second day, the Miniscope was attached and remained on for the duration of the experiment, for a total of 2âweeks. The Miniscope was connected to a lightweight coaxial cable (Cooner Wire) which connected to a low-torque passive commutator (Neurotek) to allow the mice to freely move around the home cage with minimal rotational force. After exposure to the neutral context during encoding, the mice were immediately returned to their home cage in the vivarium and the first calcium imaging recording began. The Miniscope DAQ was connected to an Arduino with a schedule set up to send a 10âmin TTL pulse to record for 10âmin, with a 20âmin break in between, repeated 24 times. Thus, we sampled 4âh worth of calcium imaging data across 12âh. The telemetry probe recorded continuously for the duration of the experiment while the mouse was in its home cage in the vivarium.
Sleep recordings and sleep scoring
The HD-X02 implants recorded EEG, EMG, temperature and locomotion continuously throughout the experiment at 100âHz. After the experiment was completed, the data were run through an automatic custom-written algorithm to detect sleep states. First, the data were binned into 6âs epochs (to allow enough cycles of slow-wave oscillations). To separate sleep and wake states, the EMG data were fit with a Gaussian mixture model with two states, in which the lower state represented sleep and the higher state represented wake. To separate REM versus NREM periods, the EEG was band-pass filtered for theta (5â9âHz) and delta (0.5â4âHz) signals, and a ratio of theta to delta signal was calculated. A Gaussian mixture model was fit to this theta/delta ratio with two states, in which high theta/delta meant REM, while low theta/delta meant NREM. The algorithm was validated against manually scored data.
Miniscope data processing and data alignment
To extract calcium transients from the calcium imaging data, we used our previously published open-source calcium imaging data processing pipeline, Minian63 v.1.2.1. In brief, videos were preprocessed for background fluorescence and sensor noise, and motion corrected. Putative cell bodies were then detected to feed into a constrained non-negative matrix factorization algorithm to decompose the three-dimensional video array into a three-dimensional array representing the spatial footprint of each cell, as well as a two-dimensional matrix representing the calcium transients of each cell. The calcium transients were then deconvolved to extract the estimated time of each calcium transient. Deconvolved calcium activities were analysed in these studies, except Extended Data Figs. 5 and 6, which used calcium traces. For calcium imaging experiments with EEG/EMG, data were processed as above; however, the videos were temporally downsampled by 2 (to 15âHz). Cells recorded across sessions within a mouse were cross-registered using a previously published open-source cross-registration algorithm, CellReg, using the spatial correlations of nearby cells to determine whether highly correlated footprints close in space are likely to be the same cell across sessions64. For calcium imaging experiments with EEG/EMG, each offline recording was cross-registered with all the encoding and recall sessions, but not with the other offline sessions because cross-registering between all sessions would lead to too many conflicts and, therefore, to no cells cross-registered across all sessions.
To align calcium imaging data with behaviour, behaviour recordings were first aligned to an idealized template assuming a perfect sampling rate. This meant that if a recording session was 5âmin, there should be 300âsâÃâ30âfpsâ=â9,000âframes (for a 30âHz recording). All behaviour recordings were within four frames of this perfect template. Calcium recordings recorded with a much more variable and dynamic sampling rate. Then, for each behaviour frame, the closest calcium imaging frame was aligned to that frame, using the computer timestamp of that frame in milliseconds. No calcium imaging frame was reused more than twice. For calcium imaging experiments with EEG/EMG, each frame of calcium activity was aligned with the sleep state the mouse was in at that time. To do this, the computer time of each calcium frame was compared with the sleep states detected around the same time. If the calcium frame occurred during one of the 6âs sleep timeframes, that calcium frame was designated that sleep state; otherwise, if there were no sleep data during that time (due to data being dropped or low quality), it was designated no state and was excluded from sleep-state-specific analyses to account for any dropped frames in the telemetry data.
General statistics and code/data availability
All analyses and statistics were performed using custom-written Python and R scripts. Code detailing all the analysis in this Article is available at GitHub (https://github.com/denisecailab/RetrospectiveMemoryLinkingAnalysis_2024). Calcium imaging data used in this Article is available through the Neurodata Without Borders framework to seamlessly share data across institutions upon reasonable request65. Statistical significance was assessed using two-tailed paired and unpaired t-tests, as well as one-way, two-way, or three-way analysis of variance, linear mixed-effects models or Ï2 tests where appropriate. Significant effects or interactions were followed with post hoc testing with the use of contrasts or with BenjaminiâHochberg corrections for multiple comparisons. Significance levels were set to αâ=â0.05. Significance for comparisons is indicated by asterisks; *Pââ¤â0.05, **Pâ<â0.01, ***Pâ<â0.001, ****Pâ<â0.0001. Sample sizes were chosen on the basis of previous similar studies. Error bars and error bands always refer to the s.e.m., and bars and points with error bars always refer to the mean. The investigators were not blinded to behavioural testing in calcium imaging studies but were blinded to behavioural testing in all other experiments. Mice were randomly assigned to groups in all of the experiments.
Ensemble reactivation analysis
To measure ensemble reactivation across the offline period (Extended Data Fig. 4f), for each mouse, the matrix of neural activity that was recorded during the offline session was z-scored along both axes (cells and time). Cells were then broken up into ensembles on the basis of whether they were previously observed to be active. Previously active cells were defined on the basis of whether they had a corresponding matched cell through CellReg. On offline 1 after neutral encoding, cells were either previously matched to an active cell during neutral encoding (neutral ensemble) or had no previously matched cell (remaining ensemble). On offline 2, cells had a matched cell only with neutral encoding and not aversive encoding (neutral ensemble), a matched cell with aversive encoding and not neutral encoding (aversive ensemble), a matched cell on both neutral encoding and aversive encoding (overlap ensemble), or no matched cell (remaining ensemble). For each ensemble, the activity of cells was averaged across cells, and then averaged across time for each time bin.
Burst participation analysis
To measure population bursts (Figs. 2 and 3 and Extended Data Figs. 3 and 4), for each mouse, all cells that were recorded during that session were z-scored along the time dimension, such that each cell was normalized to its own activity. By doing this, no cell overly contributed to population bursts by having a very high amplitude event. Then, the mean population activity across the whole population was computed across the session and that one-dimensional trace was z-scored. Time periods when the mean population activity reached above a threshold of zâ=â2 were considered to be burst events. During each of these burst events, each cell was considered to have participated if its activity was above zâ=â2 during the event. For each ensemble (as defined in the previous section), the fraction of the ensemble that participated in each event was computed, and then this was averaged across all events. The average participation of each ensemble was compared across ensembles and across low- versus high-shock groups.
Ensemble co-participation analysis
To measure ensemble co-participation during bursts (Figs. 4 and 5 and Extended Data Figs. 3, 6 and 8), bursts were defined on the basis of the z-scored mean population activity of the whole population. Then, for each burst event, the z-scored mean population activity was computed for the neutral ensemble and for the aversive ensemble (see the âEnsemble reactivation analysisâ section for ensemble definitions). For each population-level burst event, the âparticipationâ of the neutral ensemble or aversive ensemble was measured on the basis of whether the ensembleâs mean population activity was above the zâ=â2 threshold during the population level event. The burst events in which one ensemble participated without the other ensembles were considered independent participations. The burst events in which multiple ensembles simultaneously participated were considered co-participations. The fraction of burst events in which each ensemble independently participated and co-participated was computed. Then, the same computation was performed for all non-burst periods to examine how frequently the ensembles burst independently and coincidentally outside of burst events. In the calcium imaging experiment with EEG/EMG (Fig. 5), ensemble co-participation was defined above; however, as there were several offline recordings per mouse, each ensemble mean activity was computed for each offline session, and all the mean ensemble activities were concatenated to produce a pseudocontinuous time series of mean ensemble activities across the offline session. These mean activities were z-scored and then ensemble co-participation was computed separately for each sleep state.
Time-lagged cross-correlation analysis
To measure cross-correlations (Extended Data Fig. 4k), mean ensemble activities were computed for the overlap, neutral and aversive ensembles (see the previous two sections). Each time series was then broken up into 120âs bins. The overlap ensemble was separately correlated with the neutral ensemble and the aversive ensemble bin by bin. For each time bin, cross-correlations were computed for lags up to a maximum of 5 frames (or ~160âms). The maximum correlation was taken for each time bin, and the average correlation across time bins was computed. This led to, for each mouse, an average correlation between the overlap ensemble and the neutral ensemble, and an average correlation between the overlap ensemble and the aversive ensemble, across the offline period.
Inhibitory neuron chemogenetic tagging (chemotagging)
To chemogenetically identify which neurons recorded with calcium imaging were inhibitory neurons (Extended Data Fig. 6), the calcium transients of cells during the 45âmin CNO session were taken and normalized to have the range [0,1]. The number of prominent calcium peaks that each cell had from minutes 10â40 were computed and this was used to sort the cells from most to least responsive during this inhibitory tagging session (with cells with more peaks being more responsive and more likely putative GAD+ inhibitory neurons). These cells were cross-registered back to cells that were active during the previous offline 2 day (Extended Data Fig. 6b,c,h,j) to distinguish putative inhibitory neurons during that session. If a cell on offline 2 was not cross-registered with a cell on inhibitory tag day, that offline 2 cell was set to have 0 activity on inhibitory tag day, with the rationale that an hM3Dq+ cell would be likely to respond when administered with CNO. Offline cells were sorted on the basis of their responses on inhibitory tag day, with the most responsive cells being putative inhibitory neurons. Then, offline 2 cells were binned into groups on the basis of how responsive they were on inhibitory tag day (for example, top 20% of responsive cells) for downstream analyses. The same cross-registration was repeated with neutral and aversive encoding (Extended Data Fig. 6j) for decoding with putative inhibitory neurons. To compare putative inhibitory versus excitatory neurons (Extended Data Fig. 6b,c), the top 10% of most responsive cells on CNO day were used as the putative inhibitory neurons, with the rest of the population as putative non-inhibitory neurons. This is based on anatomical data estimating that inhibitory neurons make up about 10% of the neuronal population in the pyramidal layer on hippocampal CA1 (ref. 45).
SVM analyses
To perform support vector machine (SVM) decoding to distinguish neutral from aversive encoding based on neural activity (Extended Data Fig. 7a), first only cells that were active during both encoding sessions were aligned and all other cells active during only one of the encoding sessions were excluded. As neutral encoding was longer than aversive encoding, neutral encoding activity was trimmed to the same length as aversive encoding. The activity vectors were concatenated and a random 50% of vectors were used to define the training set. A linear SVM was fit to the activity patterns and then tested for decoding accuracy on the held out 50% of data. This was repeated 50 times to produce a distribution of accuracies, from which the mean accuracy was extracted. For shuffle controls, the labels were randomly shuffled and the SVM was trained on the randomly shuffled labelled data. For SVM decoding in the inhibitory tagging experiment, first decoding was done as described above (Extended Data Fig. 6i). Second, cells active during both neutral and aversive encoding were extracted, as described above. These cells were sorted on the basis of how responsive they were on inhibitory tagging day (when they received CNO). The cells were broken up into fifths from most responsive to least responsive on inhibitory tagging day. Each 20% of cells was trained using an SVM as above (Extended Data Fig. 6j). This performance was compared with shuffled label controls for each fraction of cells.
Population vector correlation analysis during encoding
To measure the similarity of population activity within and across neutral and aversive encoding, cells that were active during both neutral and aversive encoding were extracted (excluding any cells active only during one or the other), and the activities were concatenated across time. A population vector correlation matrix was computed to extract intrasession correlations (comparing every moment to every other moment within a session), as well as intersession correlations (comparing every moment within a session to all moments in the other session). The mean intrasession correlations were computed (intra-neutral and intra-aversive), as well as the intersession correlations (InterCorrs), and compared.
Encoding-to-recall population vector correlation analysis
To measure correlations between encoding and recall activity patterns (Fig. 6b), first for each mouse, only cells that were active during both the encoding and recall session were included in the analysis and were aligned across the two sessions. For the encoding session, the mean population activity across the entire session was computed to produce one vector. Then, the recall session was broken up into 30âs bins and the mean population activity vector was computed for each bin. The encoding vector was correlated with each recall vector, as described previously66. We used Kendallâs tau correlations. Finally, the correlations across all of the recall bins were averaged to produce one average correlation between encoding and recall, for each mouse.
Ensemble reactivation during neutral and novel recall
To measure reactivation of past encoding ensembles during recall (Fig. 6a and Extended Data Fig. 4qâs), for each mouse, cells active during neutral and novel recall were cross-registered with cells active during neutral encoding and not aversive encoding (neutral ensemble), aversive encoding and not neutral encoding (aversive ensemble), and during both neutral and aversive encoding (overlap ensemble). The fraction of recall cells that were cross-registered with each of these ensembles was then computed (for example, the fraction of neutral recall cells that were previously active during both neutral and aversive encodingâthe overlap ensemble, measured the reactivation of the overlap ensemble during neutral recall). These values of ensemble reactivation are reported in Extended Data Fig. 4qâs for the reactivation of the neutral, aversive and overlap ensembles during neutral and novel recall. Then, for each mouse, the difference in this reactivation between neutral and novel recall was computed (neutral reactivationââânovel reactivation) to create a reactivation index. A reactivation index of greater than 0 would indicate that an ensemble was more reactivated in neutral compared to novel recall. A value less than 0 would indicate that the ensemble was more reactivated during novel recall. These reactivation index scores are reported in Fig. 6a.
Inclusion and ethics statement
All authors support inclusive, diverse and equitable research conduct. Eight authors self-identify as part of an under-represented group in biomedical research as defined by the NIH. Moreover, nine authors, including the senior author, are women. One or more authors received support from a program designed to increase diverse representation in science, including the NIH Diversity Supplement and Mount Sinai Scholar Award.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.