Dataset
GEL is a company funded by the Department of Health and Social Care in the UK. Part of the flagship project, 100kGP, was set up to sequence 100,000 whole genomes from National Health Service (NHS) patients with rare diseases and cancer. In this study, we utilized version 12 of the cohort, comprising 14,778 participants. Sequencing libraries generated from tumour and matched germline DNA samples were sequenced using 150-base-pair paired-end reads on Illumina HiSeq platforms. In total, 16,355 tumour and 16,555 germline samples underwent whole-genome sequencing at a target depth of 100Ã for tumour and 30Ã for germline.
We included the cancer types from the following tissues (nâ=â17): breast, lung, stomach, neuroendocrine, skin, oropharyngeal, colorectal, kidney, prostate, hepato-pancreatobiliary, bladder, bone and soft tissue, ovary, endometrium, central nervous system, lymphoid and myeloid. The following tumour subtypes were then included (nâ=â39): bladder; chordoma; primary conventional osteosarcoma; liposarcoma (both dedifferentiated and myxoid); leiomyosarcoma; myxofibrosarcoma; sarcoma, not otherwise specified; HER2+ breast cancer; luminal (oestrogen receptor positive) breast cancer; triple-negative breast cancer; breast cancer, not otherwise specified; oligodendroglioma; astrocytoma; glioblastoma; adult glioma, not otherwise specified; colorectal cancer; hepato-pancreatobiliary cancer, not otherwise specified; liver hepatocellular carcinoma; cholangiocarcinoma; pancreatic adenocarcinoma; malignant pleural mesothelioma; small cell lung cancer; lung squamous cell carcinoma; lung adenocarcinoma; lung cancer, not otherwise specified; lymphoid; myeloid; neuroendocrine tumour; oropharyngeal cancer; ovarian cancer; clear cell renal carcinoma; malignant melanoma; renal cancer, not otherwise specified; upper GI squamous cell carcinoma; stomach adenocarcinoma; upper GI adenocarcinoma; upper GI cancer, not otherwise specified; and endometrial carcinoma.
Most tumour samples in the GEL cohort come from patients whose cancers were early disease stage and had not yet received treatment (Fig. 1c). Samples with tumour purity of <10% were excluded, as were cancers of unknown primary, paediatric cancers and testicular germ cell tumours (510 samples). In the cohort, 3.8% (598) of samples were fixed-formalin paraffin embedded (FFPE)30. Staging information was available for 10,780 (72.9%) patients, with 836 (5.7%) patients recorded as having stage 4 disease (Fig. 1c). Median depth of coverage for tumour samples was 97.6Ã and for germline samples was 32.6Ã. A total of 1,800 (12.1%) patients were recorded as receiving systemic anticancer treatment before biopsy. In this group, the treatment type was classified into hormonal (nâ=â27), immunotherapy (nâ=â57), targeted (nâ=â415) or cytotoxic chemotherapy (nâ=â1,653).
Inclusion and ethics
The research presented in this manuscript is compliant with ethical regulations and was approved by the East of EnglandâCambridge South Research Ethics Committee (Research Ethics Committee reference 14/EE/1112, Integrated Research Application System ID 166046). Recruitment of participants was carried out across 13 NHS Genomic Medicine Centres and all participants provided their written and informed consent.
ecDNA calls
Focal DNA copy-number alterations were identified using CNVKit v0.98. AmpliconArchitect v1.2 was used to construct cyclic paths from identified focal amplifications, and AmpliconClassifier v0.4.12 was used to determine whether these paths were likely to be ecDNA. These steps are packaged into a single workflow available at https://github.com/AmpliconSuite/AmpliconSuite-pipeline.
AmpliconArchitect identifies the structure of focal amplifications by using seed intervals that define regions that are focally amplified and extend beyond them to look for copy-number changes or discordant edges. For this analysis, seed intervals were defined as regions of greater than 50âkb, with a threshold copy number of greater than 4.5, double the ploidy of the tumour and at least 2.5 additional copies above the median arm-level copy number. The regions are then merged to form a breakpoint graph, which can be broken down into simple and complex cycles to identify any circular paths that could be indicative of ecDNA. Within the seed interval, it is possible that an ecDNA reconstruction could be less than 50âkb. AmpliconArchitect also masks highly repetitive regions such as α-satellites in centromeric and peri-centromeric regions.
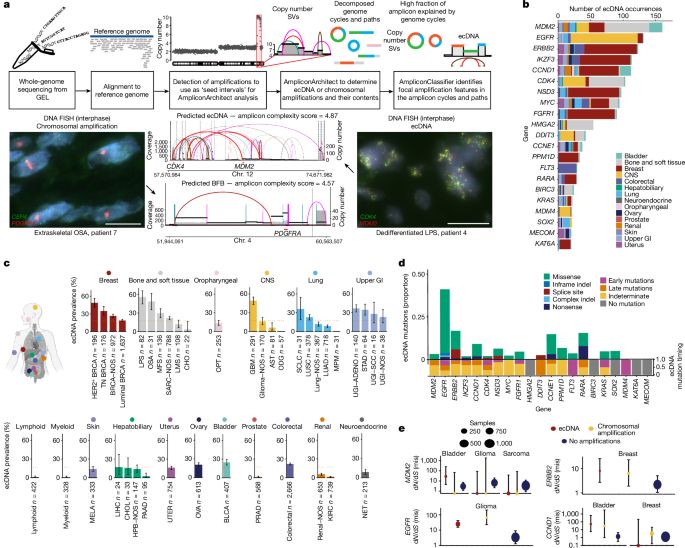
We conducted additional FISH on 11 tissue samples that we were able to obtain from GEL, demonstrating 90.9% (10/11) accuracy of our computational calls, comparable to previous validation of these methods4.
Patients were categorized as having ecDNA if ecDNA was detected in their tumour. Patients who had both chromosomal amplifications and ecDNA were included in the ecDNA category.
We then annotated each ecDNA according to whether or not it contained an oncogene as categorized by the Cancer Gene Census (https://cancer.sanger.ac.uk/census). Those that contained an oncogene were denoted as oncogenic ecDNA. We then further divided ecDNA according to whether or not any genes were annotated, classifying it as âecDNA without known oncogenesâ and âecDNA without coding genesâ. We then carried out an over-representation analysis for genes encoded on ecDNA without known oncogenes to demonstrate an enrichment for immunomodulatory genes.
Amplicon complexity
The amplicon complexity score, as defined in ref.â10, is calculated by AmpliconClassifier (Extended Data Fig. 1b). For each seed-interval-defined amplicon, AmpliconArchitect produces a copy-number-aware (CNA)-breakpoint graph. AmpliconArchitect also outputs decompositions that represent cyclic and non-cyclic paths through this CNA-breakpoint graph. These decompositions are passed as input to AmpliconClassifier to produce the complexity score, which aims to capture the diversity of the cyclic and/or non-cyclic paths present. Each path has a copy count and a length in kilobase pairs, which are combined to create a length-weighted copy number (normalized by the total length-weighted copy number present in the CNA-breakpoint graph), and the complexity score is calculated through the sum of three log-transformed measures: the total count of copy-number segments present in the amplicon; the normalized length-weighted copy number of each cyclic path; and the residual normalized length-weighted copy number that is not explained by cyclic paths.
Over-representation analysis
Over-representation analysis was carried out using the cluster profiler package (v4.6.2)31. To determine whether the genes that were annotated on âecDNA without known oncogenesâ were in predefined sets that were present at frequencies higher than expected by chance, the annotated genes were assigned to a specific gene set (denoted by the GO term). Following this, the observed proportion of genes assigned to that GO term was compared with the expected proportion given the background of all genes using Fisherâs exact test.
A false discovery rate-adjusted Pâvalue was obtained using the BenjaminiâHochberg method, with the threshold for significance set at qâ>â0.001. The minimum gene set size considered was 100. ecDNAs were considered immunomodulatory if a gene from the significant gene sets mapped to an ecDNA that did not contain an oncogene, and that significant gene set had an immunomodulatory function (GO terms: 0006968, 0002228, 0042267, 0001906, 0001909, 0002698, 0001910, 0031341, 0002367, 0002695, 0050866, 0051250, 0050777).
Permutation test for oncogene enrichment
For permutation testing, first, the proportion of focal amplifications that contained oncogenes was calculated. A total of 256 genes were identified as oncogenes. From the pool of genes identified on ecDNA at least once (nâ=â20,713), a random set of 256 genes were sampled and the proportion was calculated. This calculation was repeated 10,000 times to obtain a background distribution of the proportion of genes that belong to a gene set of equal size.
Estimates of selection using dN/dS
The dN/dS estimates were calculated using the dNdScv package16, which was run on all mutations available in the cohort. This method uses a maximum-likelihood approach in its analysis of the ratio of non-synonymous (missense, nonsense and essential splice mutations) to synonymous substitutions to infer a measure of the strength of selection acting on protein-coding genes. It also estimates a background mutation rate of each gene through joint analysis of both local and global information that takes into account sequence composition and the contribution of mutational signatures. In our analysis, estimation of dN/dS ratios was carried out across the genome, as well as stratified by context including ecDNA amplification, non-ecDNA amplification and non-amplified areas of the genome (Extended Data Fig. 10a). Six genes were amplified at >5% of the cohort with a strong signal of positive selection (dN/dS estimateâ>â5); YEATS4, CCT2, FPS6 (sarcoma), KRAS, ERBB2 (upper gastrointestinal) and EGFR (central nervous system; Extended Data Fig. 7f).
Somatic mutation calling/ploidy and purity estimation
Strelka2 (version 2.4.7)32 and Manta (version 0.28.0)33 were used to call mutations and SVs, respectively. Manta combines paired and split-read evidence for SV discovery and scoring. The following filters were applied to the raw variant calls: SVs with a normal sample depth near one or both variant break-ends three times higher than the chromosomal mean; SVs with a somatic quality score of <30; somatic deletions and duplications with a length of >10âkb; somatic small variants (<1âkb) with the fraction of reads with MAPQ0 around either break-end of >0.4. For purity estimates, CCube was used34. For ploidy estimates, the CakeTin pipeline from ref.â35 was utilized, available for 9,141 samples.
Calculation of wGII
The wGII score was calculated as the proportion of the genome with aberrant copy number relative to the median ploidy, weighted on a per chromosome basis22. Both median ploidy and copy-number segments were rounded to the nearest integer copy state from CNVKit.
SBS signature analysis
Ref.â24 quantified the fraction of SBSs found in each of the 96 trinucleotide contexts from the multiple WGS cohorts (including GEL) and analysed these data with non-negative matrix factorization to infer a set of SBS signatures24. We then used this reference set of SBS signatures to infer the most likely SBS signature for mutations in our cohort. Using the sample-level SBS exposures and the SBS reference signatures, each trinucleotide channel context is assigned a likelihood value by multiplying the sample exposure weight by the reference signature weight. This allows estimation of the most likely mutational process for each mutation.
Timing of SBS signatures
To perform this analysis we used the SBS signatures as defined in ref.â24. By analysing the variant allele frequency distribution of single nucleotide variants (SNVs) at focal amplification sites, it becomes possible to temporally assess the formation of ecDNA and the mutational processes occurring in that genomic region. This assessment involves calculating the mutational multiplicity, which is determined by the copy-number state of an SNV within a predicted ecDNA locus. SNVs are classified as occurring either pre- or post-ecDNA formation on the basis of whether the SNV copies are equivalent to the total copy number at the locus. The mutational multiplicity can be determined by the following formula:
$${\rm{C}}{\rm{P}}{\rm{N}}{\rm{m}}{\rm{u}}{\rm{t}}={\rm{V}}{\rm{A}}{\rm{F}}\times (1/p)\times (p\times {\rm{C}}{\rm{P}}{\rm{N}}{\rm{f}}{\rm{o}}{\rm{c}}{\rm{a}}{\rm{l}})+{\rm{C}}{\rm{P}}{\rm{N}}{\rm{n}}{\rm{o}}{\rm{r}}{\rm{m}}\times (1-p)$$
in which VAF represents the variant allele frequency, p represents tumour purity, CPNfocal represents the focal amplification copy number, and CPNnorm represents the local copy number in the normal genome. Mutations are considered pre-ecDNA formation if CPNmut is greater than 0.8 times CPNfocal. Mutations are classified as post-ecDNA formation if CPNmut is less than 0.8 times CPNfocal and greater than CPNnorm/2.
By aggregating mutations across multiple samples within the same tumour, a maximum-likelihood function is used to determine whether ecDNA tends to occur before or after a mutation process. This involves creating a mutational catalogue that categorizes all mutations on the basis of 96 trinucleotide context channels and their pre- or post-ecDNA formation status. Using the sample-level SBS exposures and the SBS reference signatures, each trinucleotide channel context is assigned a likelihood value by multiplying the sample exposure weight by the reference signature weight. This allows estimation of the most likely mutational process for each mutation and the identification of processes acting early or late in the context of ecDNA formation. We then carried out a Wilcoxon test, comparing the probabilities that a mutation within the ecDNA locus is early with the probability that it is late on each sample, and presented the median difference between the two categories.
Statistical analysis
All statistical tests were carried out in R (4.1.2). Correlation tests were carried out using cor.test with either Spearmanâs method or Pearsonâs method, as specified. Tests comparing distributions were carried out using wilcox.test or t.test.
Proportions were compared using prop.test. For prevalence estimates, the 95%âCI of a proportion was reported using propCI. Logistic regression models were fitted using glm(outcomeâ~âexposure_variables, family = âlogitâ), with ORs and 95%âCIs reported. For the regression analysis in which we controlled for tumour type, we excluded those tumour types with fewer than five patients sampled.
GEL sample FISH
FISH was carried out on 4âμM FFPE tissue sections according to a combination of the Agilent Technologies Protocol (Histology FISH Accessory Kit K5799) and the Abbott Molecular Diagnostic FISH probe protocol. Briefly, FFPE sections were dewaxed in xylene for 5âmin followed by rehydration in 100%, 80% and 70% ethanol and then washed twice with Agilent Technologies wash buffer. The FFPE tissue was then incubated at 98â°C for 10âmin in Agilent Technologies pretreatment solution. The Coplin jar containing the slides was removed from the 98â°C water bath and allowed to slowly cool for an additional 15âmin. The FFPE slides were washed twice with Agilent wash buffer. Stock pepsin (Agilent stock pepsin) was applied to the slide for 10âmin at 37â°C. FFPE slides were washed twice with Agilent wash buffer and then dehydrated using 70%, 80% and 100% ethanol before probe hybridization. Gene-specific probes containing chromosome-specific centromere enumeration probes (CEP) to MDM2 (+control CEP12 spectrum green) (Vysis/Abbott), MDM2 and CDK4 (Empire Genomics), PDGFRA (+control CEP4 spectrum green) (Empire Genomics) and MYC (Vysis/Abbott) were applied directly to the tissue sections with the coverslip being sealed with rubber solution glue.
Denaturation of the probes on the tissue was carried out at 75â°C for 7âmin. The slide was then incubated overnight in a moist box at 37â°C for 16âh. Slides were washed for 10âmin at 73â°C with 0.4à SSC containing 0.3% Igepal (Sigma) followed by a 10-min wash at room temperature with 2à saline-sodium citrate (SSC) containing 0.1% Igepal. Slides were allowed to air dry and then counterstained with a Vectashield mounting medium containing 4â²,6-diamidino-2-phenylindole (DAPI; ThermoFisher). Images were captured using the Applied Precision DeltaVision Microscope.
HER2 FISH
FFPE tissue sections were deparaffinized by two 5-min incubations in Histo-Clear (Electron Microscopy Sciences 64110), followed by 5âmin in 100% ethanol and 5âmin in 70% ethanol. Samples were then incubated in 0.2âN HCl for 20âmin. Slides were then placed in 10âmM antigen retrieval buffer (10âmM citric acid pHâ6.0) and incubated in a vegetable steamer (90â95â°C) for 20âmin. Slides were briefly washed in 2à SSC and then treated with proteinase K digestion buffer (1:100 dilution of proteinase K NEB P8107 in TE buffer, 100â200âμl per sample) for 1âmin at room temperature. Slides were then dehydrated by incubation for 2âmin each in 70%, 85% and 100% ethanol. The HER2 FISH and Chr.â17 control centromere enumeration FISH probes (Empire Genomics ERBB2-CHR17-20-ORGR) were diluted 1:5 in hybridization buffer (Empire Genomics), added to each slide, and covered with a coverslip. Slides were denatured at 75â°C for 5âmin followed by overnight hybridization at 37â°C in a humidified chamber. Slides were washed twice in 0.4à SSCâ+â0.3% Igepal630 (5âmin, 40â60â°C) and then in 2à SCCâ+â0.1% Igepal630 (5âmin, room temperature). Slides were treated with a TrueVIEW Autofluorescence Quenching kit (Vector laboratories SP-8400) according to the manufacturerâs directions for 2âmin and then washed in 2à SSC (5âmin, room temperature). Slides were mounted with ProLong Gold antifade with DAPI (ThermoFisher P36931). Slides were imaged on a Zeiss LSM880 confocal microscope using a 0.45-μm Z-step size. Maximum-intensity projections were generated using ZEN2.3 SP1 FP3 software. This component of the study was approved by the Stanford University Institutional Review Board (number 69198).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.