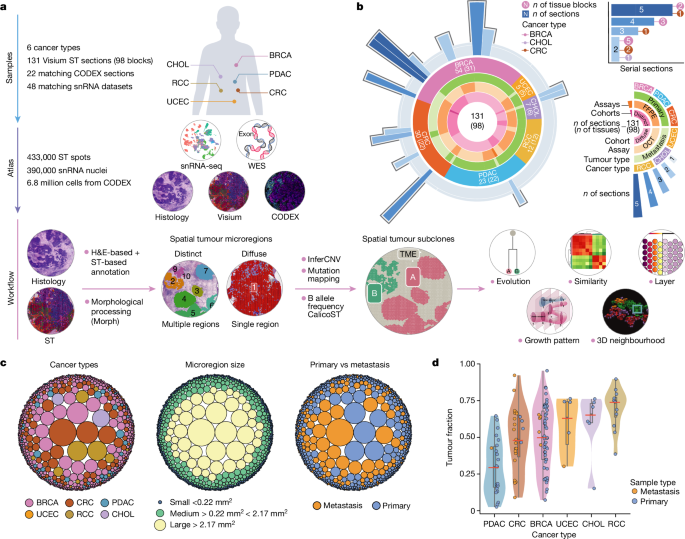
Experimental methods
Specimens and sample processing
All samples were collected with informed consent at the Washington University School of Medicine in St Louis. Samples from BRCA, PDAC, CRC, CHOL, RCC and UCEC were collected during surgical resection and verified by standard pathology (institutional review board protocols 201108117, 201411135 and 202106166). After verification, a 1.5âÃâ1.5âÃâ0.5âcm3 portion of the tumour was removed, photographed, weighed and measured. Each portion was then subdivided into 6â9 pieces and then further subdivided into 4 transverse-cut pieces. These four pieces were then each respectively placed into formalin, snap-frozen in liquid nitrogen, DMEM and snap-frozen before embedding in OCT. The purpose of choosing grid processing over punch sampling was utility-based, as it minimized remaining tissue. Relevant protocols can be found at protocols.io (https://doi.org/10.17504/protocols.io.bszynf7w)46.
ST preparation and sequencing
OCT-embedded tissue or FFPE tissue samples were sectioned and placed on a Visium Spatial Gene Expression Slide following the Visium Spatial ProtocolsâTissue Preparation guide. Samples used for serial sections were sectioned and collected with an interval range from 5 to 100âμm. When doing serial sectioning, the first section was named as U1, followed by U2, U3, and so on. Selected sections were loaded onto Visium slides and the distance between each section was recorded. For OCT-embedded samples, detailed methods have been described in a previous publication10. In brief, fresh tissue samples were coated with room temperature OCT without any bubbles. After RNA quality check using a Tapestation and a morphology check using H&E staining for the OCT-embedded tissue samples, blocks were scored into a suitable size that fit the capture areas and then sectioned into 10-μm sections. Sections were then fixed in methanol, stained with H&E and imaged at Ã20 magnification using the bright-field imaging setting on a Leica DMi8 microscope. Tissue samples were then permeabilized for 18âmin and ST libraries were constructed following the Visium Spatial Gene Expression Reagent kits user guide CG000239 Rev A (10x Genomics). cDNA was reverse transcribed from the poly-adenylated messenger RNA, which was captured using primers on the slides. Next, the second strand was synthesized and denatured from the first strand. Free cDNA was then transferred from slides to tubes for further amplification and library construction. Libraries were sequenced on a S4 flow cell of an Illumina NovaSeq-6000 system. For FFPE samples, detailed methods have been described in a previous publication47. In brief, quality control was done by evaluating DV200 of RNA extracted from FFPE tissue sections per the Qiagen RNeasy FFPE Kit protocol, then followed by performing the Tissue Adhesion Test described in the 10x Genomics protocol. Sections (5âμm) were placed on a Visium Spatial Gene Expression Slide according to the Visium Spatial ProtocolsâTissue Preparation guide (10x Genomics, CG000408 Rev A). After overnight drying, slides were incubated at 60â°C for 2âh. Deparaffinization was then performed following the protocol for Visium Spatial for FFPEâDeparaffinization, H&E staining, Imaging and Decrosslinking (10x Genomics, CG000409 Rev A). Sections were stained with H&E and imaged at Ã20 magnification using the bright-field imaging setting on a Leica DMi8 microscope. Afterwards, decrosslinking was performed immediately for H&E stained sections. Next, human whole transcriptome probe panels were added to the tissue. After these probe pairs hybridized to their target genes and ligated to one another, the ligation products were released following RNase treatment and permeabilization. The ligated probes were then hybridized to the spatially barcoded oligonucleotides on the capture area. ST libraries were generated from the probes and sequenced on a S4 flow cell of an Illumina NovaSeq 6000 system. Relevant protocols can be found at protocols.io (https://doi.org/10.17504/protocols.io.x54v9d3opg3e/v1 and https://doi.org/10.17504/protocols.io.kxygx95ezg8j/v1)48,49.
CODEX preparation and imaging
Carrier-free monoclonal or polyclonal anti-human antibodies were purchased (Supplementary Table 3) and verified using immunofluorescence (IF) staining in multiple channels. After screening, antibodies were conjugated using an Akoya Antibody Conjugation kit (Akoya Biosciences, SKU 7000009) with a barcode (Akoya Biosciences) assigned according to the IF staining results. Several common markers were directly purchased through Akoya Biosciences. CODEX staining and imaging were performed according to the manufacturerâs instructions (CODEX user manual, Rev C). In brief, 5-μm FFPE sections were placed on coverslips coated with APTES (Sigma, 440140) and baked at 60â°C overnight before deparaffinization. The next day, tissues were incubated in xylene, rehydrated in ethanol and washed in ddH2O before antigen retrieval with TE buffer, pHâ9 (Genemed, 10-0046) in boiling water for 10âmin in a rice cooker. The tissue samples were then blocked using blocking buffer (CODEX staining kit, SKU 7000008) and stained with the marker antibody panel to a volume of 200âµl for 3âh at room temperature in a humidified chamber. The dilution factor for each antibody is provided in the CODEX cycle information sheet (Supplementary Table 3). Imaging of the CODEX multicycle experiment was performed using a Keyence fluorescence microscope (model BZ-X810) equipped with a Nikon CFI Plan Apoâλ Ã20/0.75 objective, a CODEX instrument (Akoya Biosciences) and a CODEX instrument manager (Akoya Biosciences). The raw images were then stitched and processed using the CODEX processor (Akoya Biosciences). After multiplex imaging was completed, H&E staining was performed on the same tissue. Staining quality for each antibody in CODEX is shown as a single channel in green with DAPI in blue in Supplementary Figs. 10 and 11.
Single-nucleus suspension preparation
Approximately 20â30âmg of flash-frozen or cryopulverized or 200âµm of OCT sections of tissue from each sample were retrieved and aliquoted for nucleus preparation for use in a Next GEM Single Cell Multiome ATAC + Gene Expression kit or a Next GEM Single Cell 3â² Kit v.3.1 kit. Samples were resuspended in lysis buffer (10âmM Tris-HCl (pHâ7.4) (Thermo, 15567027), 10âmM NaCl (Thermo, AM9759), 3âmM MgCl2 (Thermo, AM9530G), 0.10% NP-40 substitute (% v/v) (Sigma, 74385-1L), 1âmM DTT (Sigma, 646563), 1% stock BSA solution (% v/v) (MACS, 130-091-376), nuclease-free water (Invitrogen, AM9937), plus 0.1âUâµlâ1 RNase inhibitor), resuspended and homogenized through douncing, and filtered through a 40-μm cell strainer (pluriSelect), then diluted with wash buffer (2% BSA, 1à PBS and RNase inhibitor). The filtrate was collected, then centrifuged at 500g for 6âmin at 4â°C. The nuclear pellet was then resuspended in BSA wash buffer with RNase inhibitor, stained with 7AAD, and nuclei were purified and sorted by FACS. Relevant protocols can be found at protocols.io (https://doi.org/10.17504/protocols.io.14egn7w6zv5d/v1, https://doi.org/10.17504/protocols.io.261gednx7v47/v1)50,51.
Single-cell suspension preparation
Approximately 15â100âmg of each tumour was cut into small pieces using a blade. Enzymes and reagents from a Human Tumour Dissociation kit (Miltenyi Biotec, 130-095-929) were added to the tumour tissue along with 1.75âml of DMEM. The resulting suspension was loaded into a gentleMACS C-tube (Miltenyi Biotec, 130-093-237) and subjected to the gentleMACS Octo Dissociator with Heaters (Miltenyi Biotec, 130-096-427). After 30â60âmin on the heated dissociation programme (37h_TDK_1), samples were removed from the dissociator and filtered through a 40-μm mini strainer (PluriSelect, no. 43-10040-60) or a 40-μm nylon mesh (Fisher Scientific, 22-363-547) into a 15-ml conical tube on ice. The sample was then spun down at 400g for 5âmin at 4â°C. After removing the supernatant, when a red pellet was visible, the cell pellet was resuspended using 200âμl to 3âml ACK lysis solution (Thermo Fisher, A1049201) for 1â5âmin. To quench the reaction, 10âml PBS (Corning; 21-040-CM) with 0.5% BSA (Miltenyi Biotec; 130-091-376) was added and spun down at 400g for 5âmin at 4â°C. After removing the supernatant, the cells were resuspended in 1âml PBS with 0.5% BSA, and live and dead cells were visualized using trypan blue. Finally, the sample was spun down at 400g for 5âmin at 4â°C and resuspended in 500âμl to 1âml PBS with 0.5% BSA to a final concentration of 700â1,500 cells per μl. The protocol is available at protocols.io (https://doi.org/10.17504/protocols.io.bsnqnddw)52.
Single-nucleus library preparation and sequencing
Nuclei and cells and barcoded beads were isolated in oil droplets using a 10x Genomics Chromium instrument. Single-nucleus suspensions were counted and adjusted to a range of 500â1,800 nuclei per µl using a haemocytometer. Reverse transcription was subsequently performed to incorporate cell and transcript-specific barcodes. All snRNA-seq samples were run using a Chromium Next GEM Single Cell 3â² Library and Gel Bead kit v.3.1 (10x Genomics). For the multiome kit, Chromium Next GEM Single Cell Multiome ATAC + Gene Expression was used (10x Genomics). Nuclei were then subjected to downstream protocols by 10x (Next GEM Single Cell Multiome ATAC + Gene Expression: https://cdn.10xgenomics.com/image/upload/v1666737555/support-documents/CG000338_ChromiumNextGEM_Multiome_ATAC_GEX_User_Guide_RevF.pdf. Next GEM Single Cell 3â² Kit v3.1: https://support.10xgenomics.com/single-cell-gene-expression/library-prep/doc/user-guide-chromium-single-cell-3-reagent-kits-user-guide-v31-chemistry-dual-index). Single-cell suspensions were subject to the Next GEM Single Cell 3â² Kit v.3.1 protocol. Barcoded libraries were then pooled and sequenced on an Illumina NovaSeq 6000 system with associated flow cells.
Genomic DNA extraction
Tumour tissue samples were obtained from surgically resected specimens. After a piece was removed for fresh single-cell preparation, the remaining sample was snap-frozen in liquid nitrogen and stored at â80â°C. Before bulk DNA extraction, samples were cryopulverized (Covaris) and aliquoted for bulk extraction. Genomic DNA was extracted from tissue samples with either a DNeasy Blood and Tissue kit (Qiagen, 69504) or a QIAamp DNA Mini kit (Qiagen, 51304). Genomic germline DNA was purified from cryopreserved peripheral blood mononuclear cells using a QIAamp DNA Mini kit (Qiagen, 51304) according to the manufacturerâs instructions (Qiagen). The DNA quantity was assessed by fluorometry using a Qubit dsDNA HS assay (Q32854) according to the manufacturerâs instructions (Thermo Fisher Scientific). Protocols are available at protocols.io (https://doi.org/10.17504/protocols.io.bsnhndb6)53.
WES analysis
About 100â250âng of genomic DNA was fragmented on a Covaris LE220 instrument targeting 250-bp inserts. Automated dual-indexed libraries were constructed using a KAPA Hyper library prep kit (Roche) on a SciClone NGS platform (Perkin Elmer). Up to ten libraries were pooled at an equimolar ratio by mass before the hybrid capture targeting a 5-µg library pool. The library pools were hybridized using xGen Exome Research Panel v.1.0 reagent (IDT Technologies), which spans a 39-Mb target region (19,396 genes) of the human genome. The libraries were hybridized for 16â18âh at 65â°C followed by a stringent wash to remove spuriously hybridized library fragments. Enriched library fragments were eluted and PCR cycle optimization was performed to prevent overamplification. The enriched libraries were amplified using KAPA HiFi master mix (Roche) before sequencing. The concentration of each captured library pool was determined through qPCR using a KAPA library Quantification kit according to the manufacturerâs protocol (Roche) to produce cluster counts appropriate for the Illumina NovaSeq-6000 instrument. Next, 2âÃâ150 paired-end reads were generated targeting 12âGb of sequence to achieve around 100à coverage per library.
Xenium library preparation and imaging
Paraffin blocks (FFPE blocks) were sectioned at 5âμm and placed on Xenium slides following the FFPE Tissue Preparation guide (10x Genomics, CG000578, Rev B). Those slides underwent a series of xylene and ethanol washes for deparaffinization and decrosslinking, using the FFPE tissue enhancer as outlined (10x Genomics, CG000580, Rev B). Overnight in situ probe hybridization was performed using 379 probes from the Xenium Human Multi-Tissue Panel (10x Genomics, 1000626) plus an additional 100 custom probes (Supplementary Table 6). After hybridization probes were ligated, the sample underwent rolling circle amplification, and the background was quenched using an autofluorescence mixture. Nuclei were stained with DAPI to improve sample tracking and approximate cell boundaries (10x Genomics, CG000582, Rev D). These samples, along with buffers and decoding consumables, were loaded into a Xenium analyzer (10x Genomics, 1000481). The run was initialized using the guidance provided (10x Genomics, CG000584, Rev C). These fluorescent reporters hybridized to targeted complementary regions of the barcoded circularized cDNA were imaged. H&E staining was performed on the same region after the run was complete.
Analytical methods
Quantification and statistical analysis
All data analyses were conducted in R and Python environments. Details of specific functions and libraries are provided in the relevant methods sections above. Significance was determined using the Wilcoxon rank-sum test, proportion test, hypergeometric test or Pearson correlation test, as appropriate. Pâvaluesâ<â0.05 were considered significant. Details of statistical tests are provided in the figure legends and the relevant methods sections.
WES data processing
FASTQ files were preprocessed using trimGalore (v.0.6.7; with parameters: –length 36 and all other parameters set to default; https://github.com/FelixKrueger/TrimGalore). FASTQ files were then aligned to the GDCâs GRCh38 human reference genome (GRCh38.d1.vd1) using BWA-mem (v.0.7.17) with parameter -M and all others set to default. The output SAM file was converted to a BAM file using the samtools (https://github.com/samtools/samtools; v.1.14) view with parameters -Shb, and all others set to default. BAM files were sorted and duplicates were marked using Picard (v.2.6.26) SortSam tool with the following parameters: CREATE_INDEX=true, SORT_ORDER=coordinate, VALIDATION_STRINGENCY=STRICT, and all others set to default; and MarkDuplicates with parameter REMOVE_DUPLICATES=true, and all others set to default. The final BAM files were then indexed using the samtools (v.1.14) index with all parameters set to default.
Mutation calling using WES
Somatic mutations were called from WES data using the Somaticwrapper pipeline (v.2.2; https://github.com/ding-lab/somaticwrapper), which includes four different callers: Strelka (v.2.9.10)54, MUTECT (v.1.1.7)55, VarScan (v.2.3.8)56 and Pindel (v.0.2.5)57. We kept exonic single nucleotide variants (SNVs) called by any two callers among MUTECT (v.1.1.7), VarScan (v.2.3.8) and Strelka (v.2.9.10) and insertions and deletions (indels) called by any two callers among VarScan (v.2.3.8), Strelka (v.2.9.10) and Pindel (v.0.2.5). For the merged SNVs and indels, we applied a 14Ã and 8Ã minimal coverage cut-off for tumour and normal tissue, respectively. We also filtered SNVs and indels by a minimal VAF of 0.05 in tumours and a maximal VAF of 0.02 in normal samples. We also filtered any SNV within 10âbp of an indel found in the same tumour sample. Finally, we rescued the rare mutations with VAFs within 0.015 and 0.05 based on an established gene consensus list58,59. In a downstream step, we used Somaticwrapper to combine adjacent SNVs into double-nucleotide polymorphisms using COCOON (https://github.com/ding-lab/COCOONS), as reported in a previous study60.
Mutation mapping to snRNA-seq and ST data
We applied an in-house tool called scVarScan that can identify reads supporting the reference allele and variant allele covering the variant site in each cell by tracing cell and molecular barcode information in a snRNA-seq and single-cell RNA sequencing (scRNA-seq) or Visium bam file. The tool is freely available at GitHub (https://github.com/ding-lab/10Xmapping). For mapping, we used high-confidence somatic mutations from WES data produced by Somaticwrapper (described above). Visium reads were prefiltered with the flag âxf:i:25â for reads contributing to unique molecular identifier counts.
Spatial mutation VAF statistical test
For each ST section, we applied two sets of statistical tests to all WES-based somatic mutations mapped to ST. First, for each mutation with greater than 30 reads of coverage on ST across all spots, the VAF was calculated for all tumour region spots and all non-tumour region spots as the number of variant reads across all spots divided by the number of total reads across all spots. A binomial test was then done using VAF of non-tumour spots as the background: binom.test(alterative=âgreaterâ). Then, a proportion test was done between the VAFs in different spatial subclones with prop.test(alternative=âtwo.sidedâ). Finally, multiple testing correction was done on both sets of tests with the function p.adjust().
CNV calling using WES
Somatic CNVs were called using GATK (v.4.1.9.0)61. Specifically, the hg38 human reference genome (NCI GDC data portal) was binned into target intervals using the PreprocessIntervals function, with the bin length set to 1,000âbp and using the interval-merging-rule of OVERLAPPING_ONLY. A panel of normals was then generated using each normal sample as input and the GATK functions CollectReadCounts with the argument –interval-merging-rule OVERLAPPING_ONLY, followed by CreateReadCountPanelOfNormals with the argument –minimum-interval-median-percentile 5.0. For tumour samples, reads that overlapped the target interval were counted using the GATK function CollectReadCounts. Tumour read counts were then standardized and denoised using the GATK function DenoiseReadCounts, with the panel of normals specified by –count-panel-of-normals. Allelic counts for tumours were generated for variants present in the af-only-gnomad.hg38.vcf according to GATK best practices (variants further filtered to 0.2 > af > 0.01 and entries marked with âPASSâ) using the GATK function CollectAllelicCounts. Segments were then modelled using the GATK function ModelSegments, with the denoised copy ratio and tumour allelic counts used as inputs. Copy ratios for segments were then called on the segment regions using the GATK function CallCopyRatioSegments.
Bedtools62 intersection was used to map copy number ratios from segments to genes and to assign the called amplifications or deletions. For genes overlapping multiple segments, a custom Python script was used to call that gene as amplified, neutral or deleted based on a weighted copy number ratio calculated from the copy ratios of each overlapped segment, the lengths of the overlaps and the zâscore threshold used by the CallCopyRatioSegments function. If the resulting zâscore cut-off value was within the range of the default zâscore thresholds used by CallCopyRatioSegments (v.0.9,1.1), then the bounds of the default zâscore threshold were used instead (replicating the logic of the CallCopyRatioSegments function).
ST data processing
For each sample, we obtained the unfiltered featureâbarcode matrix per sample by passing the demultiplexed FASTQ files and associated H&E image to Space Ranger (v.1.3.0, v.2.0.0 and v2.1.0 âcountâ command using default parameters with reorient-images enabled) and the prebuilt GRCh38 genome reference 2020-A (GRCh38 and Ensembl 98). Seurat was used for all subsequent analyses. We constructed a Seurat object using the Load10X_Spatial function for every slide. Each slide was then scaled and normalized with the SCTransform function to correct for batch effects. Any merged analysis or subsequent subsetting of cells and samples for a sample with several slides underwent the same scaling and normalization method. Spots were clustered using the original Louvain algorithm, and the top 30 principal component analysis dimensions using the FindNeighbors and FindClusters functions as described in the âAnalysis, visualization, and integration of spatial datasets with Seuratâ vignette from Seurat (https://satijalab.org/seurat/articles/spatial_vignette.html).
InferCNV and CalicoST for CNV calling on Visium ST data
To detect large-scale chromosomal CNVs using scRNA-seq, snRNA-seq and Visium data, InferCNV (v.1.10.1) was used with default parameters recommended for 10x Genomics data (https://github.com/broadinstitute/inferCNV). InferCNV was run at the sample level and only with post-quality control filtered data using the raw counts matrix. For snRNA-seq and scRNA-seq data, all non-malignant cells were used as a reference with the annotation ânon-tumourâ and all malignant cells had the same annotation âtumourâ, with the following parameters: analysis_mode=âsubclustersâ, –cluster_by_groups=T, –denoise=T, and –HMM=T. For Visium ST data, 200 spots annotated as ânon-malignantâ with the lowest ESTIMATE purity score were used as a reference, and âmalignantâ spots had their microregion ID as annotation, with the following parameters: window_length=151, analysis_mode=âsampleâ, –cluster_by_groups=T, –denoise=T, and –HMM=T. CalicoST (https://github.com/raphael-group/CalicoST)63 was run on Visium ST data with the same input annotation (microregion ID). All spots from the same microregions were treated as the smallest unit of analysis. CalicoST was then run with default parameters with results manually inspected.
Copy number profile similarity score calculation
To determine the similarity between two spatial CNV profiles, we use a modified Jaccard similarity score. A CNV profile was defined as a set of genomic windows with annotation copy number neutral (0), amplification (1) or deletion (â1). Two CNV profiles were then compared, and overlapping genomic windows were broken down so that both profiles had the same sets of windows (with the function reduced from the package GenomicRanges v.1.46.1). Then, the CNV similarity score (Sim) was defined as follows:
$${{\rm{Sim}}}_{A,B}=\frac{\sum _{i}{\rm{size}}\left({w}_{i}\right)\times ({{\rm{CNV}}}_{A,i}\times {{\rm{CNV}}}_{B,i})}{\sum _{i}{\rm{size}}\left({w}_{i}\right)}$$
where \({w}_{i}\) denotes the size of the genomic window i, \({{\rm{CNV}}}_{A,i}\) denotes the CNV annotation (0, 1 or â1) for profileâA in genomic window i, and \({{\rm{CNV}}}_{B,i}\) denotes the CNV annotation for profileâB in genomic window i across all genomic windows where either A or B is not CNV neutral.
To determine the similarity between a spatial CNV profile and WES-based CNV (related to Extended Data Fig. 5a), we used a similarity score averaging the sensitivity (fraction of WES-based CNVs also detected in spatial CNVs) and specificity (fraction of spatial CNVs agreeing with WES-based CNVs). Specifically,
$${{\rm{Sim}}}_{A,E}=\left(\frac{\sum _{a}{w}_{a}\times ({{\rm{CNV}}}_{A,a}\times {{\rm{CNV}}}_{E,a})}{\sum _{a}{w}_{a}}+\frac{\sum _{e}{w}_{e}\times ({{\rm{CNV}}}_{A,e}\times {{\rm{CNV}}}_{E,e})}{\sum _{e}{w}_{e}}\right)/2$$
where \({w}_{a}\) denotes the size of the genomic window a from spatial CNV, \({w}_{e}\) denotes the size of the genomic window e from a WES-based CNV, \({{CNV}}_{A,a}\) denotes the CNV annotation (0, 1 or â1) for profileâA in genomic window a.
Spatial subclone identification based on CNV profile similarity
In the OCT workflow (Supplementary Fig. 1a), CalicoST simultaneously identified CNVs and groups microregions into spatial subclones. In the FFPE workflow, confident spatial CNV events in each microregion were first selected by comparing them with matching WES. Then, a pairwise CNV similarity score was calculated across all tumour microregions. Finally, microregions were clustered with CNV similarity scores using the function hclust (dâ=â1-CNV similarity, method=âward.D2â), and divided into clusters with function cutree (hâ=â0.8âÃâmax(hclust$height)). Final subclone assignments were manually reviewed to avoid overclustering and to eliminate small outlier CNV profiles.
Tumour microregion annotation and layer determination
Using Visium ST, tumour microregions were determined through a multistep process using H&E. Each ST spot was assigned as either stroma or tumour by manually reviewing the morphology on H&E stained sections. If at least 50% of the pixels within a spot covered malignant cell morphology, the spot was labelled as tumour. Otherwise, it was labelled as stroma. Next, we defined distinct tumour microregions using a set of three rules. The first rule specified that tumour spots immediately adjacent to one another are initially marked as a single tumour microregion. The second rule states that if two distinct tumour regions together occupied at least 50% of one single spot, the spot is assigned to the distinct tumour region with the higher percentage occupied. Finally, the third rule specified that if there was a clear morphological difference of the tumour spots within one tumour microregion, the microregion must be separated into distinct microregions, one per clear morphology.
Afterwards, we ran the Morph toolset (https://github.com/ding-lab/morph), which uses mathematical morphology to refine the tumour microregions. That is, if the total number of spots in a microregion is less than or equal to three, then we labelled all such spots as stroma. Last, Morph assigned the layer (for example, T1) of each spot of a tumour microregion by a sequence of mathematical morphology operations described in the Spot-depth correlation analysis method, which denotes the depth of a given spot inside a microregion.
Average spot area and microregion size calculation
To calculate the area each spot takes, we used the spot size (55âµm) and centre-to-centre distance between each spot (100âµm) provided by 10x Genomics (http://kb.10xgenomics.com/hc/en-us/articles/360035487572-What-is-the-spatial-resolution-and-configuration-of-the-capture-area-of-the-Visium-v1-Gene-Expression-Slide-). As illustrated in Supplementary Fig. 6, the Visium spots form a hexagonal lattice that covers the sample. The repeating unit of this lattice is a trapezoid shape centred at each spotâs centre that is composed of eight equilateral triangles. Each triangle has a side of 50âµm (half of the spot the centre-to-centre distance). Using the area equation of equilateral triangles and multiplying it by 8, we obtained the area of each trapezoid as 8,660âµm2, which is the average area occupied by each spot. To calculate the microregion size, we multiplied the spot count by 8,660 and divided by 106 to obtain the size in mm2.
Micoregion density estimation
We estimated microregion density per section by following the formula: density per µm2â=ân microregion per section size (in spots) then divided by 8,660âµm per spot. Then density per mm2â=âdensity per µm2âÃâ106 (n microregion per mm2).
Cell-type annotation
Cell-type assignment was done based on the following known markers: Bâcell, CD79A, CD79B, CD19, MS4A1, IGHD, CD22 and CD52; cDC1, CADM1, XCR1, CLEC9A, RAB32 and C1orf54; cDC2, CD1C, FCER1A, CLEC10A and CD1E; mregDC, LAMP3, CCR7, FSCN1, CD83 and CCL22; pDC, IL3RA, BCL11A, CLEC4C and NRP1; macrophage, CX3CR1, CD80, CD86, CD163 and MSR1; mast cell, HPGD, TPSB2, HDC, SLC18A2, CPA3 and SLC8A3; endothelial, EMCN, FLT1, PECAM1, VWF, PTPRB, ACTA2 and ANGPT2; fibroblast, COL1A1, COL3A1, COL5A1, LUM and MMP2; pericyte, RGS5, PLXDC1, FN1 and MCAM; NK cell, FCGR3A, GZMA and NCAM1; plasma cell, CD38, SDC1, IGHG1, IGKC and MZB1; Tâcell, IL7R, CD4, CD8A, CD8B, CD3G, CD3D and CD3E; and regulatory Tâcell, IL2RA, CTLA4, FOXP3, TNFRSF18 and IKZF2. Normal epithelial cells in the breast were annotated with the following markers: LumSec, GABRP, ELF5, CL28, KRT15, BARX2 and HS3ST4; LumHR, ANKRD30A, ERBB4, AFF3, TTC6, ESR1, NEK10 and XBP1; and basal, SAMD5, FBXO32, TP63, RBBP8 and KLHL13. Normal epithelial cells in the liver were annotated with the following markers: hepatocyte, ALB, CYP3A7, HMGCS1, ACSS2 and AKR1C1; cholangiocyte, SOX9, CFTR and PKD2. Normal epithelial cells in the pancreas, including ductal, acinar, islet-α, islet-β and islet-γ cells, were annotated with singleR (v.1.8.1) using reference data BaronPancreasData(âhumanâ).
Spot-depth correlation analysis
We identified a correlation between gene expression and spot depth in its tumour microregion. First, each spot was assigned a depth defined as the distance to the closest TME-facing spot in its tumour microregion. This depth was quantified in several layers through an iterative process whereby all the malignant spots immediately adjacent to non-malignant spots were considered layerâ1, and then all malignant spots immediately adjacent to layerâ1 were considered layer 2, and the process was repeated until all spots were assigned with a layer number. If a spotâs layer was larger than the smallest distance between the spot and any Visium border (including the edge of the Visium capture window, edge of the tissue section and any empty spots inside the section), then we excluded such spots, as we only knew the upper bound of the depth of this spot. Additionally, tumour microregions with fewer than 3âlayers or 50âspots were excluded from the analysis. The distance between layers was taken as the centre-to-centre distance of Visium spots (100âµm).
To give the same weight to bigger and smaller regions, the depth of each spot was further normalized by the maximum depth of the microregion this spot belonged. Then, we performed partial correlation tests independently between gene expression (at least 1 transcript detected from the gene in more than 50% of all spots) and normalized depth of each spot, with tumour purity as a covariate as follows:
$${\rm{Expression}}={\rm{rho}}\times ({\rm{layer}}\;{\rm{fraction}})+b\times {\rm{purity}}$$
where layer fraction is the layer number divided by the total number of layers in a tumour to normalize for large and small microregions, rho is the layer correlation coefficient, and b is the correlation coefficient for covariant purity. Purity was inferred with deconvolution when there was matching snRNA-seq data (deconvoluted tumour fraction per spot by RCTD), or with ESTIMATE (that is, tumour purity estimate score per spot) otherwise. Each gene was checked against a set of snRNA-seq-derived non-malignant gene lists to ensure that the change in fraction did not derive from a shift in cell type composition. Finally, we performed multiple-testing adjustments for all tests done in each ST section.
Spot-depth GSEA pathway enrichment analysis
To summarize biological programs enriched in the centre and periphery of tumour microregions across sections, we first obtained the cohort-level average layer correlation coefficient. If a test was not significant (Pââ¥â0.05), rho was assigned to be 0 to indicate no correlation. If a test was not performed on a section (<50% of the spots have at least one transcript), rho was also assigned as 0. When a case had multiple sections, we first took the average rho across sections to avoid bias towards tumours with more sections. Then, the average of rho was calculated for each cohort (all samples or samples from each cancer type).
In the same fashion, rank statistics were calculated for each test as âlog10(Pâvalue)âÃârho for tests with Pâ<â0.05. For tests with Pââ¥â0.05 or genes not tested, the rank statistic was 0. We then calculated average rank statistics per case, followed by the average per cancer type. Finally, with the full list of rank statistics calculated for all genes tested, we used the function GSEA (parameters: pvalueCutoff=0.5; package: clusterProfiler v.3.18.1) to obtain the normalized enrichment score of Hallmark pathways (package: msigdbr 7.5.1) from the MSigDB64. Finally, only pathways with Pâ<â0.1 were kept in the final results.
Tumour intrinsic and non-tumour gene categorization
We use differential expression and perâcent expression filters, comparing expression among cell types in the matching snRNA-seq data to further characterize genes identified in the centre and periphery enriched analysis. The steps implemented in this workflow generated four categories: tumour-specific, stromal-specific, tumour-enriched and stromal-enriched (Supplementary Fig. 8a,b). Genes that did not pass the significant cutoff in any differential expression analysis were labelled separately as not DEG.
To distinguish these four groups, we first performed differential gene analysis of cell types in the matching snRNA-seq data, filtered by a conventional significance cut-off (log2(fold change)â>â0.5, adjusted Pâ<â0.05, Bonferroni correction), to obtain DEGs (Supplementary Fig. 8a). Given the heterogeneity in tumours, certain tumour-specific genes might only exist in a subpopulation of tumours. Therefore, we first subclustered the tumour populations (using the Subcluster function in Seurat with a resolution of 0.5) to obtain tumour subclusters. We then compared each subcluster with all other non-tumour cells. A gene was considered a tumour DEG if at least one tumour subcluster showed significant expression compared with the non-tumour cells and vice versa for non-tumour DEGs (Supplementary Fig. 8a,b).
For candidate tumour or stromal-specific genes, a DEG was designated as tumour-specific if it met both of the following criteria: (1) it is a DEG when compared with all non-tumour cell types from at least one tumour subcluster; and (2) its expression was <15% in all non-tumour cell types (Supplementary Fig. 8c).
The reverse applied to candidate stromal-specific DEGs. If a DEG did not meet both of these requirements to be tumour or stromal specific, it was designated as either tumour-enriched or stromal-enriched based on whether the expression level was higher in tumour or stromal cell types (Supplementary Fig. 8a).
Spatial subclone-specific treatment response analysis
We focused on ten cases (comprising four BRCA, two CRC and four PDAC samples) with multiple spatial subclones for this analysis. To obtain subclone-specific DEGs, we used FindMarkers from the function in Seurat with the âwilcoxâ test option DEGs between each subclone and TME. We then applied the cut-off for adjusted Pâ<â0.01, average log2(fold change)â>â1 and perâcent expression in at least one cell typeâ>â0.4 to select significant DEGs. To infer treatment response, we used the perturbation database LINCS L1000 (ref. 65), specifically the LINCS_L1000_Chem_Pert_down dataset from Enrichr66, to evaluate the gene set overlap between upregulated DEGs in spatial subclones and downregulated genes after compound treatment. To make the plot in Supplementary Fig. 4, we sorted the data by âOdd.Ratioâ and selected top compounds from each subclone. The corresponding compound metadata, including mechanism of action, was obtained from CLUE (clue.io, âExpanded CMap LINCS Resource 2020 Releaseâ) to add annotation on the heatmap.
Organ-specific gene blacklist for non-malignant cell types
To distinguish transcripts originating from cancerous versus non-malignant stromal or immune cells, we used merged snRNA-seq data per organ (breast, kidney, liver and pancreas) for cell-type marker analysis. This analysis used the FindAllMarkers function in Seurat with the âwilcoxâ test option. Subsequently, we refined the gene list by applying filters such as average log2(fold change)â>â2, perâcent expression in at least one cell typeâ>â0.4 and adjusted Pâvaluesâ<â0.01 to ensure robust marker selection for each cell type. The resultant gene list is available in Supplementary Table 5. This list was instrumental in excluding non-cancerous cell genes from analyses pertaining to cancer-specific expression patterns, such as pairwise microregion similarity analysis. Of note, during the analysis, we observed a notable mapping of various epithelial cell types in the snRNA-seq reference dataset for BRCA when using the RCTD deconvolution method. This observation probably stems from the diverse BRCA subtypes present in the cohort. To address this, we opted to combine all epithelial cell types into a single category during the identification of cell-type markers and excluded them from the blacklist. For tumours originating from organs other than the four mentioned above, we aggregated all genes present in the blacklist across organs to form a comprehensive multiorgan blacklist, which aided in filtering out non-cancerous transcripts.
Microregion transcriptional profile analysis
For overall tumour heterogeneity, we selected Morph-identified spots then ran ROGUE (v.1.0)67 to measure heterogeneity as 1-ROGUE. We then compared the transcriptional profiles of microregions by selecting the top 500 most variable features after excluding stroma regions in ST samples following Morph processing. Our initial evaluation involved conducting Pearson correlation tests for each pair of microregions, using a range of the top 250â1,500 most variable genes with increments of 250 (that is, 250, 500, 750, â¦, 1,500). We observed consistent correlations for nearly all values beyond using more than 500, which led us to select the top 500 genes for this analysis. This choice reduced the risk of selecting too few variable genes (for example, <250 most variable genes) while also avoiding the inclusion of numerous genes with minimal effect on the transcriptional profile. GSEA analysis was done using the function GSEA (parameters: pvalueCutoffâ=â0.5; package: clusterProfiler v.3.18.1) to obtain the normalized enrichment score of Hallmark pathways (package: msigdbr v.7.5.1) from the MSigDB64.
Module score calculation
Module scores on top of each heatmap in Extended Data Fig. 6 were calculated with the AddModuleScore function from Seurat68 using the genes listed in each heatmap. This score represents the average expression levels of a gene set. The score was calculated for each spot and a box plot was used to show the distribution of module scores in each microregion.
ST cell-type decomposition
Cell-type composition per spot was deconvolved using RCTD18 with default parameters and doublet_mode = âmultiâ. The reference for each run was the cell types manually annotated from the Seurat object of the matching snRNA-seq or Multiome sample. To quantify spatial distribution of each cell type, cell type fraction of 6 layers (T3 and above, T2, T1, E1, E2, E3 and above) from each tumour microregion is calculated and averaged in each sample. To compare differential TME infiltration between spatial subclones, cell type fraction from all spots between spatial subclones was compared with pairwise Wilcoxon rank-sum test and FDR adjustment.
Spatial cellâcell interaction at tumour boundary
We evaluated the spatial-based cellâcell interaction (CCI) in the ST sample using COMMOT69 with CellChat database and distance threshold of 1,000âµm, following the same threshold used in the original publication for Visium. The median sender and receiver signals for each interaction family were compared between all tumour boundary spots (including tumour boundary layer and TME boundary layer) and all non-boundary spots (Wilcoxon rank-sum test) on a sample. Interaction pathways with signal difference great than 0.1 and FDR less than 0.05 are considered significantly boundary-enriched. Boundary DEGs were identified with FindMarkers function on three sets of comparisons: boundary/tumour, boundary/TME and boundary/all non-boundary. A boundary DEG has adjusted P value 0.25 in boundary/non-boundary test, and log2(fold change) > 0 in the other two tests.
Serial section alignment and branching factor calculation
We applied PASTE2 (ref. 70), the updated ST-based alignment tool PASTE70, to enable partial image alignment. Serial sections of the same tumour piece were aligned pairwise with default settings. Each Visium data point in every ST section received new coordinates, denoted as xâ² and yâ², based on the alignment results. We then identified the nearest spot on each adjacent section for every spot, connecting them along the zâaxis. This process facilitated the linking of spots across all sections on the zâaxis. To assess whether one microregion was connected to another in an adjacent section, we first removed stromal spots and then counted the connected spots. If any microregion on one section connected to the next section with more than three shared spots, then we considered these two microregions, located on different sections, as connected in 3D space and forming the same tumour volume. This connection was labelled as volumeâ1, volumeâ2, and so forth in the figures (Fig. 5d,e and Extended Data Fig. 9aâd).
We used two geometric metrics to describe tumour volume: connectivity and loop. For connectivity (degree), this metric quantifies the number of connections from an individual microregion to adjacent sections. For example, if microregionâ2 in sectionâ2 connects to 3 microregions in sectionâ1 and 2 in sectionâ3, its connectivity is 5. The maximum connectivity of a tumour volume is the highest connectivity among its microregions. For loop, this metric was calculated as the total number of connections minus the total number of microregions plus one, identifying intricate loop structures within the tumour volume.
Registration of Visium, CODEX and H&E serial sections
Before registration, imaging data underwent the following transformations. Multiplex images were converted to greyscale images of DAPI intensity. The image was then downscaled by a factor of 5 before key point selection. H&E images (also downsampled by a factor of 5) were used for keypoint selection with Visium data.
For registration, we used BigWarp71, which was packaged in the Fiji/ImageJ software application. To register each collection of serial images, we used the first serial section as the fixed image and the second image as the moving image. After the second image was warped to the first image, the second image was used as the fixed image for the transformation of the third image. Key point registration proceeded in this fashion for all images in the serial section experiment. A total of 4â20 key points were selected per image transformation. Once key points were selected, a moving field was exported from BigWarp for each image transformation. This dense displacement field was then upscaled by a factor of 5 so it could be used to warp the full-resolution imaging data. The full-resolution dense displacement field was then used to register its corresponding multiplex or Visium data. The code used for registration is available at GitHub (https://github.com/ding-lab/mushroom/tree/subclone_submission).
Neighbourhood identification input preprocessing
Once imaging data were registered, they were processed in the following manner before model input.
For Visium ST data, genes were limited to genes expressed in a minimum of 5% of spots across all serial sections and expression counts were log2 transformed. CODEX, Visium ST and H&E data were normalized by subtracting the mean expression and dividing by the standard deviation for each gene.
Expression profiles for each patch were generated differently for image-native data (CODEX and H&E) and point-based data (Visium). Expressions for CODEX and H&E patches were calculated as the average pixel intensities for each image channel over all pixels within the patch bounds. Visium patches were calculated in largely the same manner; however, the expression profile of each spot within the patch was linearly weighted by its distance to the centre of the patch. This differential weight helped to account for variation expression due to the number of spots that fall within patch boundaries.
Neighbourhood identification model architecture
The neighbourhood annotation model consisted of an autoencoder with a vision transformer (ViT) backbone (Supplementary Fig. 7). In brief, an autoencoder is an unsupervised training method for which an encoder (embedding component) and a decoder (reconstruction component) work together to learn how input data are generated. Specifically, the network derives an approximation, Q, to the true posterior generating function, P, for the output, given the input. The autoencoder used was asymmetric, meaning that the encoder and decoder were not inverse copies of one another. The encoder consisted of a ViT with a similar architecture to previously described architectures72,73 (Supplementary Table 4).
ViTs work on image tokens as input. In brief, image tokens are n-dimensional representations of patches of the input image. During training, image tiles were sampled from a uniform distribution across the set of input sections (Supplementary Fig. 7a). The sampled tile was then split into patches, for which the number of patches was determined by two hyperparameters: patch height (ph) and patch width (pw). Each patch was then flattened to a 1âÃâ(phâÃâpwâÃâc) vector, where c is the number of channels in the image (in the case of spatial transcriptomics data, c is the number of genes). The unrolled patches were then concatenated into a nâÃâ(phâÃâpwâÃâc) matrix, where n is the number of patches in the image tile. Each row in this matrix is a token that represents a patch in the image tile. The tokens were then projected by a linear layer to shape nâÃâd, where d is the dimension of the transformer blocks.
After this, a slide token was concatenated to patch tokens. The slide token (representing the slide from which the image tile was selected) was indexed from a trainable embedding of size n_slidesâÃâd, where n_slides is the number of slides in the serial section experiment. The motivation for the slide token is that as it is passed through the transformer blocks, along with the patch tokens, information can be shared across all tokens, allowing the slide token to learn to attend to useful representations of the patches. This feature allowed the model to be more robust to batch effects between serial sections. Following the addition of the slide token, positional embeddings were added to all tokens and passed through the transformer blocks comprising the ViT encoder. All variables above and details of the transformer architecture are available in Supplementary Table 4.
Once passed through the encoder, patches were represented as an embedding of size nâÃâd. The next step of the architecture was neighbourhood assignment. Neighbourhoods were assigned to patches in a hierarchical manner, meaning that a patch was classified into several neighbourhoods that differed in level of specificity. For each level of the neighbourhood hierarchy, the subsequent levels comprised partitions of the previous levelsâ neighbourhoods, that is, except for the first level, each neighbourhood was a subset of a neighbourhood in a previous level of the hierarchy. For this analysis, the model generated three levels of neighbourhoods, each with the capacity to discover up to nâ=â8 (levelâ1), nâ=â32 (levelâ2) and nâ=â64 (levelâ3) neighbourhoods, respectively. For this analysis, all neighbourhoods shown are neighbourhoods annotated at hierarchy levelâ3. The model contained three codebooks (one for each level) that are of size n_NBHDsâÃâd, where n_NBHDs is the number of possible neighbourhoods that can be assigned for the given level. The patch embeddings output by the ViT encoder were projected by three independent blocks of linear layers (one for each level) that output each patchâs probability of assignment to a given neighbourhood. These probabilities were then used to retrieve neighbourhood embeddings from the codebook corresponding to the neighbourhood level. Three linear blocks (one for each level) were then used to independently reconstruct patch embeddings at each level to each patchâs original pixel values. The code used for training the model is available at GitHub (https://github.com/ding-lab/mushroom/tree/subclone_submission).
Model loss function
The overall loss function has two main contributions: mean squared error (MSE) on the reconstruction of the input patches, and cross-entropy loss on the encoded distribution and the normal distribution with 0 mean and 1.0 variance.
During training, the autoencoder was simultaneously trying to optimize two main tasks: the reconstruction of the expression profile of each image patch embedding and the alignment of neighbourhood labels between adjacent sections. These two competing objectives forced the model to learn representative expression patterns while also keeping neighbourhoods aligned between input sections, which helped to combat neighbourhood differences due to batch effects. Differences in patch expression were quantified by MSE, whereas neighbourhood adjacency was enforced by minimizing the cross-entropy of patches adjacent to each other in the zâdirection during training.
The overall loss function is defined below:
$${{\bf{L}}}_{{\rm{o}}{\rm{v}}{\rm{e}}{\rm{r}}{\rm{a}}{\rm{l}}{\rm{l}}}={\lambda }_{{\rm{N}}{\rm{B}}{\rm{H}}{\rm{D}}}{{\bf{L}}}_{{\rm{N}}{\rm{B}}{\rm{H}}{\rm{D}}}+{\lambda }_{{\rm{M}}{\rm{S}}{\rm{E}}}{{\bf{L}}}_{{\rm{M}}{\rm{S}}{\rm{E}}}$$
Where λNBHD (maximum of 0.01) and λMSE (set to 1.0) are scalers for the neighbourhood loss (LNBHD) and reconstruction loss (LMSE), respectively. During training, λNBHD for was linearly increased from 0 to its maximum value.
Model training and inference
Two separate runs of the model were trained for HT397B1 (six H&E, four CODEX and two Visium ST slides) and HT268B1 (four Visium ST slides). Training hyperparameters, such as batch size and number of training steps, are provided in Supplementary Table 4. For HT268B1, only one instance was trained because only one data type was present. For HT397B1, three model instances were trained (one for each data type) and were subsequently merged following the procedure described in the section â3D neighbourhood construction and integrationâ.
Following training, the model inference was performed on overlapping image tiles for each slide using a sliding window of size 8 and a stride of 2 (that is, 2 overlapping patches between image tiles). The 2âÃâ2 centre patches of each tile were extracted and retiled to match the original slide orientation. Each reconstructed âpatch embedding imageâ was at a resolution of 50âpixelsâµmâ1 (that is, each neighbourhood patch represents an area that is 50âµm wide) with the exception of Visium ST, for which the patch resolution was 100âpixelsâµmâ1.
3D neighbourhood construction and integration
After the assignment of neighbourhoods for each section, slides were interpolated to generate a 3D neighbourhood volume. For this, we used linear interpolation of neighbourhood assignment probabilities with the torchio library74.
Following interpolation, we also integrated neighbourhood volumes for HT397B1, for which multiple data-type-specific volumes were generated using a graph-based clustering approach. In brief, all overlapping neighbourhood voxel annotations were identified. A graph was then constructed, whereby nodes represented each neighbourhood partition combination, and edges are the distance (in the expression profile) between these partition combinations. This graph was then clustered with the Leiden graph clustering algorithm to identify integrated neighbourhoods. Hyperparameters for the above clustering process are provided in Supplementary Table 4. 3D neighbourhoods were displayed using the open-source visualization tool Napari (https://github.com/napari/napari).
Analysis and quantification of 3D neighbourhoods
Neighbourhoods were then assigned to Visium ST spots in the following manner. Each spot was assigned the neighbourhood label of the neighbourhood overlapping its spot centroid.
To focus on neighbourhoods most related to the TME biology, we filtered out neighbourhoods with >50% overlap with copy number annotated subclones. Additionally, we excluded neighbourhoods that mapped to fewer than ten total spots across all ST sections for a sample.
The subclone boundary region for tumour clones was defined as the union of the outermost layer of subclone annotated spots and the spots one layer expanded out from them, representing an area roughly 100â150âµm at the tumourâTME interface. Subclone-specific fractions were calculated as the neighbourhood overlaps with the outermost layer of each subclone.
In HT397B1, DEGs were calculated for all neighbourhoods, not only those filtered for subclone overlap and spot count. The top 50 DEGs for neighbourhoodsâ4 and 6 were grouped into three categories: shared, unique to neighbourhoodâ4 and unique to neighbourhoodâ6. For the display in Fig. 5, the top 10 for each group were selected for display based on the following sorting criteria. The mean expression delta between neighbourhoodsâ4 and 6 was calculated for each gene by subtracting the mean expression in neighbourhood 6 from neighbourhoodâ4. Shared DEGs were ordered in ascending fashion based on the absolute expression delta of each gene. Genes unique to neighbourhoodâ4 and neighbourhoodâ6 were ordered by mean expression delta in descending and ascending fashion, respectively.
Cell-type annotation of CODEX imaging data
Our workflow for cell annotation consisted of four main steps: (1) image format conversion, (2) cell segmentation, (3) spatial feature generation and (4) cell-type classification. First, we converted image output by the CODEX platform (.qptiff) to the popular open-source OME-TIFF format. During this process, we also produced a separate image for each sample, as multiple sections of tissue are sometimes included on the same imaging run. We then used the Mesmer pre-trained nucleiâ+âmembrane segmentation model in the DeepCell framework75 to segment nuclei and whole cells. DAPI was used as the nuclei intensity image, and the channels pan-cytokeratin, HLA-DR, SMA, CD4, CD45, Hep-Par-1, CD31, E-cadherin, CD68 and CD3e were, for those present in a given image, mean-averaged to a single channel and used as the membrane intensity image.
We then use a gating procedure to identify cell types. First, to combat differences in protein intensity distributions between imaging runs and tissue types, thresholds were manually set for all protein channels used during cell typing for each image by visual inspection. Above this intensity threshold, a pixel was considered positive for a given marker, and below it, a pixel was considered negative. We then used the cell segmentation boundaries from the previous step to calculate the fraction of positive pixels for all cell typing markers in each cell. The result of this process is a feature matrix (num cellsâÃânum proteins) representing positive marker fractions for each cell typing protein in every cell. A cell was considered positive for a marker if >5% of its pixels were positive for that marker. Cells were then labelled with a gating strategy specific to each sample. During gating, each cell was subjected to a series of AND gates, whereby if a cell passed all criteria for a given step, it was annotated as the cell type specified for that step, whereas if it failed the criteria it was passed on to the next downstream step in the gating strategy. The gating strategies used for the samples in this paper are presented in Supplementary Table 4.
The following labels were the set of all possible cell type annotations: epithelial, CD4 Tâcell, CD8 Tâcell, regulatory Tâcell, Tâcell, macrophage, macrophage-M2, Bâcell, dendritic, immune, endothelial, fibroblast and hepatocyte. For some images, not all proteins required to gate a specific cell type were present. For example, CD4 was not in every image panel and available to use in the annotation of CD4 Tâcells. In these instances, the gating strategy was constructed such that cells can be labelled as more general cell types if specific proteins are not present (that is, labelled more broadly as Tâcell instead of CD4 Tâcell). If a cell was negative for all steps in the gating strategy, it was annotated as âunlabelledâ. The code for image format conversion and cell segmentation can be found at GitHub (https://github.com/estorrs/multiplex-imaging-pipeline).
Distance to tumour boundary quantification on CODEX
After registration, Visium spots labelled as tumours were mapped to CODEX slides using the coordinates of the aligned images. The coordinates of the centre of each spot in the CODEX-aligned slide were the same as its Visium counterpart. Each spot in the CODEX-aligned slide occupied the area of a circle with a radius of 150âpixels. The Euclidean distance transforms in the CODEX-aligned slide were then calculated for each pixel using Pythonâs scipy.ndimage.distance_transform_edt. Both the distances from the microregions and within the microregions were calculated.
3D tumour volume reconstruction and location quantification
The surface mesh visualizations of the tumour volumes for HT397B1 and HT268B1 were generated using the following steps: (1) tumour neighbourhood selection, (2) mesh construction and (3) mesh colouring. First, integrated neighbourhoods with tumour metrics (described below) exceeding a given threshold were considered to be part of the tumour volume. In HT268B1, the metric used to quantify epithelial character was the fraction of subclone annotated Visium ST spots per neighbourhood. Neighbourhoods with >60% subclone spots were considered tumour. In HT397B1, we instead used the fraction of CODEX-annotated epithelial cells, as CODEX sections outnumbered the Visium ST sections for that sample. Neighbourhoods with >60% fraction of epithelial cells were considered tumour.
A new volume was then constructed whereby neighbourhoods classified as tumour neighbourhoods using the above criteria were considered tumour-positive voxels, and all other voxels were tumour-negative. This 3D tumour mask was then smoothed with a Gaussian kernel (sigmaâ=â1.0). The resulting values were then used as input for the marching cubes algorithm76,77 to generate a surface mesh for the tumour volume. We used the scikit-image implementation (skimage.measure.marching_cubes) of the marching cubes algorithm with default parameters.
To colour the surface mesh, we generated 3D feature volumes (described below), and then coloured points on the surface mesh based on the voxel value at the corresponding location in the feature volume. A feature volume is a volume whereby each voxel in the volume describes some feature from the serial section dataset (for example, expression of a given gene, fraction of cells, and so on). Feature volumes used in this analysis were constructed in the following manner. First, in the serial sections for which a feature was applicable, the feature was binned at the same resolution as the 3D neighbourhoods (50âµm in this case). The binned feature was then interpolated in the zâdirection to fill in gaps between sections. The resulting volume was of the same shape as the integrated neighbourhood volume, for which the value of each voxel was the aggregated feature count for the voxel. For HT268B1, the features used were logged expression of TYMP1 and IGLC2. For HT397B1, we used fibroblast and immune cell fraction. Cells were annotated as described in the section âCell-type annotation of CODEX imaging dataâ. The surface mesh was visualized using Napari (https://github.com/napari/napari) and contrast was adjusted on a volume-to-volume basis. We also visualized the HT397B1 tissue volume with the Imaris platform, for which we generated surfaces from the following CODEX markers: pan-cytokeratin (epithelial), CD45 (immune) and SMA (stromal).
Xenium probe design
Custom Xenium gene and mutation probes were designed using Xenium Panel Designer (https://cloud.10xgenomics.com/xenium-panel-designer) following instructions outlined in the âGetting Started with Xenium Panel Designâ instructions (https://www.10xgenomics.com/support/in-situ-gene-expression/documentation/steps/panel-design/xenium-panel-getting-started#design-tool). In brief, 21-bp sequences flanking the targeted transcribed variant site were curated from the Ensembl canonical transcript (Ensembl v.100). All four possible ligation junctions (two for the WT allele and two for the variant allele, three in the case of deletionsâtwo for the WT allele and one for the variant allele) were then evaluated. Variant sites for which only non-preferred junctions (CG, GT, GG and GC) were available were excluded. The two bases of the ligation junction sequence were the last base of the RBD5 (RNA binding domain) and the first base of the RBD3 probe. Preferred junctions were always prioritized over neutral junctions unless a neutral junction was necessary to avoid hairpins, homopolymer regions, dimers or an unfavourable annealing temperature. Probe lengths for RBD5 and RBD3 were then adjusted from the 21-bp starting length to target a temperature between 50â°C and 70â°C per probe (overall target 68â°C and 82â°C). Variant sites with probes predicted to form dimers or hairpins by IDTâs oligo analyzer were excluded. Variant sites with homopolymer regions of five consecutive bases or more in either the RBD5 or RBD3 probes were excluded.
Spatial expression deconvolution
Here we used both deconvolution results and cell-type-specific expressions in the snRNA-seq data to deconvolve the Visium ST expression data (Supplementary Fig. 9). In brief, for a given Gene1, we first calculated the average expression of Gene1 per cell type in matched snRNA-seq data, subsequently filtering out the expression of such genes in cell types having <5% of the highest average expression, and then dividing each cell-type average expression from the sum of all average expressions, thereby creating the expression contribution per cell type matrix (Q). Then for a given spot, the contribution per cell type was multiplied by cell type proportion from the cell type devolution result (for example, RCTD), then normalized to 1 to give a final expression contribution matrix (WN). For instance, in Supplementary Fig. 9a, Gene1 has 40%, 30% and 30% contributions from respective cell types A, B and C based on the filtered snRNA-seq expression. For Spot1, as there is only 1 cell type, B, in the spot, 40%âÃâ1/40%âÃâ1 gives the final 100% contribution of Gene1 to cell type B in Spot1. Spot2 contains 50% A and B cell types, respectively, the normalized cell type contribution in spot 2 is therefore 50%âÃâ40%/(50%âÃâ40%â+â50%âÃâ30%)âââ57.1% for the cell typeâA, and 50%â Ãâ30%/(50%âÃâ40%â+â50%âÃâ30%)âââ42.9% for the cell typeâB. The final deconvolved expression was obtained by multiplying the original expression per spot (5 and 20 in Spot1 and Spot2) with the respective cell-type-based contribution to obtain the final deconvoluted expression values of Spot1âââcell typeâBâ=â5, Spot2âââcell typeâAâââ10.42, and Spot2âââcell typeâBâââ8.58.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.