Mice
All animal experiments were approved by the Institutional Animal Care and Use Committee at Rockefeller University and were carried out in accordance with the National Institutes of Health guidelines. Mice were group housed in a 12-h light/12-h dark cycle at 22â°C and 30â60% humidity with ad libitum access to a normal chow diet and water. We used the following mouse genotypes: C57BL/6J (wild type (WT); stock no. 000664, The Jackson Laboratory), NPY-IRES2-FlpO-D (The Jackson Laboratory, stock no. 030211), Rosa26-LSL-Cas9 (The Jackson Laboratory, stock no. 024857) and BNC2-P2A-iCre (see below). For all Cre or Flp mouse line experiments, only heterozygous animals were used. Sample sizes were decided on the basis of experiments from similar studies. Littermates of the same sex were assigned randomly to either experimental or control groups. The experiments were performed using both male and female mice, as indicated.
Generation of BNC2-P2A-iCre mouse line
The BNC2-P2A-iCre mouse line was generated by the CRISPR and Genome Editing Resource Center and Transgenic and Reproductive Technology Resource Center at Rockefeller University using CRISPRâCas9 technology51. Briefly, a custom-designed long single-stranded DNA (lssDNA) donor, containing homology arms of Bnc2 locus flanking the P2A-iCre sequence, was inserted near the endogenous STOP codon. Two guide RNAs (sgRNAs) were used to induce site-specific double-stranded breaks. The lssDNA donor with the pre-assembled Cas9 proteinâgRNA complexes was mixed and microinjected into C57BL/6J mouse zygotes following standard CRISPR genome engineering protocols. The resulting live offspring were genotyped by PCR with two sets of primers that specifically amplified the mutant allele. Validation was ensured by Sanger sequencing. The BNC2-P2A-iCre transgenic animals were bred to C57BL/6J mice for maintenance.
Viruses
AAVs used in these studies were obtained from Addgene, UNC Vector Core, or generated by Janelia Viral Tools Service. We used the following viruses: AAV5-hSyn-DIO-hM3D(Gq)-mCherry (Addgene, catalogue no. 44361, 2.2âÃâ1013âvgâmlâ1), AAV5-hSyn-DIO-hM4Di(Gi)-mCherry (Addgene, catalogue no. 44362, 2.5âÃâ1013âvgâmlâ1), AAV5-hSyn-DIO-mCherry (Addgene, catalogue no. 50459, 2.2âÃâ1013âvgâmlâ1), AAV5-Ef1a-DIO-EYFP (Addgene, catalogue no. 27056, 1.6âÃâ1013âvgâmlâ1), AAV5-hSyn-Flex-GCaMP6s-WPRE (Addgene, catalogue no. 100845, 2.9âÃâ1013âvgâmlâ1), AAV5-EF1a-DIO-hChR2(H134R)-EYFP (UNC Vector Core, 2.7âÃâ1012âvgâmlâ1), AAV1-hSyn1-SIO-stGtACR2-FusionRed (Addgene, catalogue no. 105677, 2.1âÃâ1013âvgâmlâ1), AAV5-Ef1a-fDIO-mCherry (Addgene, catalogue no. 114471, 2.3âÃâ1013âvgâmlâ1), AAV8-Ef1a-fDIO-GCaMP6s (Addgene, catalogue no. 105714, 2.1âÃâ1013 vgâmlâ1), AAV5&DJ-EF1a-fDIO-hChR2(H134R)-EYFP-WPRE (UNC Vector Core, 1.4âÃâ1012âvgâmlâ1), AAV5-Ef1a-mCherry-flex-dtA (Addgene, catalogue no. 58536, 3.88âÃâ1012âvgâmlâ1). For Lepr deletion, AAV viral vectors were cloned inhouse and packaged with the AAV5 serotype using Janelia Viral Tools Service. The sequences of sgLepR are: 5â²-GAGTCATCGGTTGTGTTCGG-3â², 5â²-TGCCGGCGGTTGGATG GACT-3â² (virus titre, 4.9âÃâ1012âvgâmlâ1); The sequence of sgCtrl is: 5â²-TTTTTTTTTTTTTTGAATTC-3â² (virus titre, 8.5âÃâ1012âvgâmlâ1). Viral aliquots were stored at â80â°C before stereotaxic injection.
Chemicals and diet
The following chemicals were used in this study: Leptin (ThermoFisher Scientific, catalogue no. 498OB05M, 3âmgâkgâ1 or 5âmgâkgâ1, intraperitoneal injection), Sema (Millipore Sigma, catalogue no. AT35750, 10ânmolâkgâ1, subcutaneous injection), CNO dihydrochloride (Tocris, catalogue no. 6329, 3âmgâkgâ1, intraperitoneal injection), sucrose tablets 20âmg (TestDiet, catalogue no. 1811555) and HFD (Research Diets, 60% kcal% Fat).
Stereotactic surgery
Mice (8â10âweeks old) were anaesthetized using isoflurane anaesthesia (induction 5%, maintenance 1.5â2%) and positioned on a stereotaxic rig (Kopf Instruments, Model 1900). Viruses were delivered into the brains through a glass capillary using a Drummond Scientific Nanoject III Programmable Nanoliter Injector. For the ARC region, the following coordinates relative to the bregma were used: anteriorâposterior, â1.65âmm to â1.70âmm; medialâlateral (ML), ±0.25âmm to 0.30âmm and dorsalâventral (DV), â5.9âmm. For chemogenetics experiments, Bnc2 neuron labeling and Lepr deletion, 30â50ânl of the virus was injected bilaterally at a rate of 1ânlâsâ1. For optogenetics, 30ânl of the virus was injected unilaterally at a rate of 1ânlâsâ1 followed by the implant of an optical fibre (ThorLabs, catalogue no. CFM12U-20) at 200âµm above the ARC (anteriorâposterior, â1.65âmm; ML, 0.3âmm; DV, â5.7âmm). For fibre photometry experiments, 30ânl of the virus was injected unilaterally followed by the implant of an optical fibre cannula (Doric, catalogue no. MFP_400/430/1100-0.57_1m_FCM-MF2.5_LAF) at 150âµm above the ARC (anteriorâposterior, â1.65âmm; ML, 0.3âmm; DV, â5.75âmm). For CRACM experiments, the two viruses were mixed at the ratio of 1:1, and 50ânl of the mixed virus was injected bilaterally into the ARC.
Isolation of nuclei and snRNA-seq
Male C57BL/6J mice aged 10â12 weeks were euthanized by transcardial perfusion using ice-cold HEPES-Sucrose Cutting Solution containing NaCl (110âmM), HEPES (10âmM), glucose (25âmM), sucrose (75âmM), MgCl2 (7.5âmM) and KCl (2.5âmM) at pHâ7.4 (ref. 52). Subsequently, brains were dissected quickly in the same solution, frozen using liquid nitrogen and stored at â80â°C until nuclei isolation. To isolate nuclei, as described previously53,54, the samples were thawed on ice, resuspended in HD buffer containing tricine KOH (10âmM), KCl (25âmM), MgCl2 (5âmM), sucrose (250âmM), 0.1% Triton X-100, SuperRNaseIn (0.5âUâmlâ1), RNase Inhibitor (0.5âUâmlâ1). Samples were homogenized using a 1âml dounce homogenizer. The resulting homogenates were filtered using a 40âμM filter, centrifuged at 600g for 10âmin and resuspended in nucleus storage buffer containing sucrose (166.5âmM), MgCl2 (10âmM), Tris buffer (pHâ8.0, 10âmM), SuperRNaseIn (0.05âUâmlâ1), RNase Inhibitor (0.05âUâmlâ1) for subsequent staining. Nucleus quality and number were assessed using an automated cell counter (Countess II, ThermoFisher). For staining, nuclei were labelled with Hoechst 33342 (ThermoFisher Scientific, catalogue no. H3570; 0.5âµl per million nuclei), anti-NeuN Alexa Fluor 647-conjugated antibody (Abcam, catalogue no. ab190565; 0.5âµl per million nuclei) and TotalSeq anti-Nuclear Pore Complex Proteins Hashtag antibody (BioLegend, catalogue no. 682205; 0.5âmg per million nuclei) for 15âmin at 4â°C. Following staining, samples were washed with 10âml 2% BSA (in PBS) and centrifuged at 600g for 5âmin. Nuclei were then resuspended in 2% BSA (in PBS) with RNase inhibitors (SuperRNaseIn 0.5âUâmlâ1, RNase Inhibitor 0.5âUâmlâ1) for subsequent fluorescence-activated cell sorting. The samples were gated on the basis of Hoechst fluorescence to identify nuclei and then further sorted on the basis of high Alexa Fluor 647 expression, designating NeuN+ nuclei as neurons.
Nuclei were captured and barcoded using 10x Genomics Chromium v.3 following the manufacturerâs protocol. The processing and library preparation were carried out by the Genomics Resource Center at Rockefeller University, and sequencing was performed by Genewiz using Illumina sequencers.
SnRNA-seq analysis
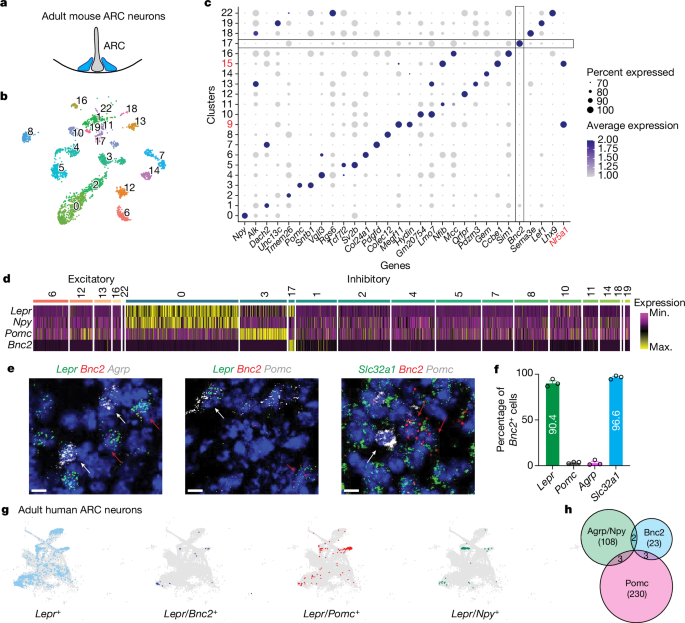
The FASTQ file was analysed with Cell Ranger v.5.0. The snRNA-seq data for ARC (WT) was preprocessed individually using the Seurat v.4 (v.4.0.3)55. Cells with more than 800 and fewer than 5,000 RNA features were selected for further analysis. Cells with greater than 1% mitochondrial genes and greater than 12,000 total RNA counts were also removed. Genes detected in fewer than three cells were excluded. We then demultiplexed the cells on the basis of their hashtag count (positive quantileâ=â0.8) using the built-in function in Seurat v.4. Only the cells with singlet Hashtag assignment were kept for downstream analysis. The data was then log-normalized with a scale factor of 10,000. After the initial quality control, demultiplexing and normalization steps, all the singlets were then scaled and reduced dimensionally with principal component analysis and uniform manifold approximation and projection (UMAP). Leiden clustering (resolutionâ=â0.55) was applied to identify clusters. We used known cell-type specific gene expression to annotate the clusters.
We analysed co-expression of marker genes within the human ARC using previously published human adult samples, and the data can be accessed through the NeMO archive (https://assets.nemoarchive.org/dat-917e9vs). A cell was considered to express the marker gene if at least two unique molecular identifiers were detected. The identification of arcuate cells was achieved by clustering and the expression of canonical markers, as detailed in the earlier study. Co-expression of genes such as Lepr, Bnc2, Agrp, Npy and Pomc was tabulated in R, and two-tailed Fisherâs tests were calculated to assess the significance of co-expression of gene pairs within the 16,819 arcuate cells in the human dataset.
Chemogenetics for activation or inhibition
AAV viruses were delivered bilaterally into the ARC of male BNC2-Cre mice aged 8â10âweeks. Mice were then allowed to recover and express DREADDs for at least 3âweeks. For activation or inhibition, animals were injected intraperitoneally with 3âmgâkgâ1 of CNO or PBS (control).
Optogenetics for activation or inhibition
AAV viruses were delivered unilaterally into the ARC of male BNC2-Cre mice aged 8â10âweeks followed by the implantation of an optic fibre. Subsequently, the mice were given a recovery period of at least 3âweeks to allow for gene expression. Before the experiments, the mice were habituated to patch cables over a period of 5âdays. The implanted optic fibres were connected to patch cables using ceramic sleeves (Thorlabs) and linked to a 473ânm laser (OEM Lasers/OptoEngine). The output of the laser was verified at the beginning of each experiment. A blue light, generated by a 473ânm laser diode (OEM Lasers/OptoEngine) with a power of 15âmW, was used. The light pulse (10âms) and frequency (20âHz) were controlled by a waveform generator (Keysight) to either activate or inhibit BNC2 neurons in the ARC. In the activation feeding experiments, mice were allowed to acclimate to the cage for 20âmin. Subsequently, three feeding sessions, each lasting 20âmin, were initiated. During these sessions, the light was turned off for the initial 20âmin, switched on for the subsequent 20âmin and then turned off again for the remaining 20âmin. In the inhibition feeding experiments, following the 20âmin acclimation, each feeding session was extended to 30âmin. The amount of food consumed during each feeding session was recorded manually. Animals were euthanized at the end of the experiments to confirm viral expression and fibre placement using immunohistochemistry.
Real-time place preference
A custom-made two-chamber box (50âÃâ50âÃâ25âcm black plexiglass) with an 8.5âcm gap enabling animals to move freely between the chambers was used for this assay. To evaluate the initial preference of the mice, they were introduced into the box for a 10âmin session without any photostimulation. Subsequently, in the second 10âmin session following the initial one, photostimulation (15âmW, 20âHz) was paired with the chamber for which the mice exhibited less preference during the initial session. The Ethovision XT v.13 software, coupled with a CCD camera, facilitated the recording of the percentage of time spent by the mice in each chamber.
Fibre photometry
Mice were acclimated to tethering and a home-cage-style arena for 5âmin daily over the course of 5âdays before the experiment. Data acquisition was conducted using a fibre photometry system from Tucker-Davis Technologies (catalogue no. RZ5P, Synapse) and Doric components, with recordings synchronized to video data in Ethovision by transistorâtransistor logic triggering. A dual fluorescence Mini Cube (Doric) combined light from 465ânm and isosbestic 405ânm light-emitting diodes (LEDs), which were transmitted through the recording fibre connected to the implant. GCaMP6s fluorescence, representing the calcium-dependent signal (525ânm), and isosbestic control (430ânm) were detected using femtowatt photoreceivers (Newport, catalogue no. 2151) and a lock-in amplifier at a sampling rate of 1âkHz. Analysis was conducted using a Matlab script involving the removal of bleaching and movement artifacts using a polynomial least square fit applied to the 405ânm signal, adjusting it to the 465ânm trace (405fitted), and then calculating the GCaMP signal as %ÎF/Fâ=â(465signalâââ405fitted)/405fitted. The resulting traces were filtered using a moving average filter and down-sampled by a factor of 20. The code is available upon request.
In situ hybridization
Mice were briefly transcardially perfused with ice-cold RNase-free PBS. Brains were then quickly collected, embedded in optimal cutting temperature embedding medium on dry ice, and stored at â80â°C until cryostat sectioning (15âµm thickness) onto Superfrost Plus Adhesion Slides (ThermoFisher). The RNAscope Fluorescent Multiplex assay (Advanced Cell Diagnostics Bio) was based on the manufacturerâs protocol. All reagents were purchased from Advanced Cell Diagnostics (ACDbio). Probes for the following mRNAs were used: Agrp (catalogue no. 400711-C3), Pomc (catalogue no. 314081-C3), Lepr (catalogue no. 402731), Slc31a1 (catalogue no. 319191) and Bnc2 (catalogue no. 518521-C2). Briefly, brain sections were fixed in 4% paraformaldehyde (PFA) at 4â°C for 15âmin followed by serial submersion in 50% ethanol, 70% ethanol, and twice in 100% ethanol for 5âmin each at room temperature. Sections were treated with ProteaseâIV for 30âmin at room temperature followed by a 2âh incubation with specific probes at 40â°C using a HyBez oven. Signal amplification was achieved through successive incubations with Amp-1, Amp-2, Amp-3 and Amp-4 for 30, 15, 30 and 15âmin, respectively, at 40â°C using a HyBez oven. Each incubation step was followed by two 2âmin washes in RNAscope washing buffer. Nucleic acids were counterstained with DAPI Fluoromount-G (SouthernBiotech) mounting medium before coverslipping. The slides were visualized using an inverted Zeiss LSM 780 laser scanning confocal microscope with a Ã20 lens. The acquired images were imported into Fiji for further analysis.
Immunohistochemistry
Mice were perfused transcardially with PBS first and then 4% PFA for fixation. Brains were collected and immersed in 4% PFA overnight at 4â°C for more fixation. Fixed brains were immersed sequentially in 10% sucrose, 20% sucrose and 30% sucrose buffers for 1âh, 1âh and overnight, respectively, all at 4â°C. After this, the brains were embedded in optimal cutting temperature embedding medium and stored at â80â°C until cryostat sectioning (30â50âµm thickness). For the staining process, brain sections were first blocked in a blocking buffer containing 3% BSA, 2% goat serum and 0.1% Triton X-100 in PBS for 30âmin at room temperature followed by an overnight incubation with primary antibodies in the cold room. After washing in PBS, the sections were incubated with fluorescence-conjugated secondary antibodies (Invitrogen) for 1âh at room temperature. Stained sections were mounted onto SuperFrost (Fisher Scientific catalogue no. 22-034- 980) slides and then visualized with an inverted Zeiss LSM 780 laser scanning confocal microscope with a Ã10 or Ã20 lens. The acquired images were imported to Fiji for further analysis. The following antibodies were used: FOS antibody (1:1,000; Synaptic systems, catalogue no. 226308), pSTAT3 antibody (1:1,000; Cell Signaling Technology, catalogue no. 9145âs), GFP (1:1,000; abcam, catalogue no. ab13970), RFP (1:1,000; Rockland, catalogue no. 600-401-379).
Electrophysiology and CRACM
Adult mice were euthanized by transcardial perfusion using ice-cold cutting solution containing choline chloride (110âmM), NaHCO3 (25âmM), KCl (2.5âmM), MgCl2 (7âmM), CaCl2 (0.5âmM), NaH2PO4 (1.25âmM), glucose (25âmM), ascorbic acid (11.6âmM) and pyruvic acid (3.1âmM). Subsequently, brains were quickly dissected in the same solution and sectioned using a vibratome into 275âµm coronal sections. These sections were then incubated in artificial cerebral spinal fluid containing NaCl (125âmM), KCl (2.5âmM), NaH2PO4 (1.25âmM), NaHCO3 (25âmM), MgCl2 (1âmM), CaCl2 (2âmM) and glucose (11âmM) at 34â°C for 30âmin, followed by room temperature incubation until use. The intracellular solution for current-clamp recordings contained K-gluconate (145âmM), MgCl2 (2âmM), Na2ATP (2âmM), HEPES (10âmM) and EGTA (0.2âmM, 286âmOsm, pHâ7.2). The intracellular solution for the voltage-clamp recording contained CsMeSO3 (135âmM), HEPES (10âmM), EGTA (1âmM), QX-314 (chloride salt, 3.3âmM), Mg-ATP (4âmM), Na-GTP (0.3âmM) and sodium phosphocreatine (8âmM, pHâ7.3 adjusted with CsOH). Signals were acquired using the MultiClamp 700B amplifier and digitized at 20âkHz using DigiData1550B (Molecular Devices). The recorded electrophysiological data were analysed using Clampfit (Molecular Devices) and MATLAB (Mathworks).
For CRACM experiments, voltage-clamp recordings were conducted on BNC2 and NPY neurons. To record oIPSCs, the cell membrane potential was held at 0âmV. ChR2-expressing axons were activated using brief pulses of full-field illumination (0.5âms, 0.1âHz, ten times) onto the recorded cell with a blue LED light (pE-300 white; CoolLED). Subsequently, TTX (1âµM), 4-AP (100âmM) and PTX (1âµM) were applied sequentially through the bath solution, each for 10â20âmin. Data acquisition started at least 5âmin after each drug application.
Indirect calorimetry
Indirect calorimetry was performed using the Phenomaster automated home cage phenotyping system (TSE Systems). Mice were housed individually in environmentally controlled chambers maintained at 22â°C, following a 12âh light/12âh dark cycle, and at 40% humidity, with ad libitum access to food and water. O2 and CO2 measurements were collected at 15âmin intervals with a settling time of 3âmin and a sample flow rate of 0.25âlâminâ1. The raw data obtained were analysed using CalR56.
Blood glucose, GTT and ITT
Blood glucose levels were measured using a OneTouch Ultra meter and glucose test strips. For GTTs, mice were fasted overnight followed by a 20% glucose injection (2âgâkgâ1) and glucose measurements at 0, 15, 30, 60 and 120âmin. ITTs were conducted after a 4âh fast, with insulin injection (0.75âUâkgâ1) and glucose measurements at 0, 15, 30, 45 and 60âmin. To test how BNC2 neuron activity affected glucose metabolism, CNO was injected for 1âh before the start of GTT and ITT experiments.
Statistical analysis
All statistical analyses used GraphPad Prism v.9. Data distribution was tested for normality (ShapiroâWilk test) and then comparisons were made using parametric or non-parametric tests, as appropriate. Two-tailed statistical tests were used, and statistical significance was determined by Studentâs t-test, MannâWhitney test, Fisherâs exact test, one-way or 2-way ANOVA, and Friedman test as indicated in the Source Data.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.