Design of DNA bricks and templates
The DNA templates (T960, T1200, L1, L2, L3, L4 and L5) and bricks were designed by a computer-assisted method. First, the reported software40 was used to generate a series of DNA brick sequences. By using the design software, a set of short DNA bricks with a length of 24ânt was generated without a four-repeat base domain, while keeping the hamming distance at least four and the content of CG between 40% and 60%. Second, we analysed the DNA bricks and templates manually to test the DNA sequences to avoid complex secondary structures and mismatches between bricks and templates (Supplementary Fig. 6).
Fluorescence assay
All DNA fluorescent experiments were performed in 1à Tris-acetate-EDTA (TAE)/Mg2+ buffer at 25â°C using real-time fluorescence PCR (CFX Connect Real-Time System, Bio-Rad). DNA b1 (or b2) was labelled with fluorophore (FAM) at 5â² ends and a1 (or a2) was labelled with quencher (BHQ-1) at 3â² ends. The FAM fluorescence signal was detected at 492ânm excitation and 518ânm emission. In a typical reaction, 30âμl solution was used for the detection. The time dependence of the fluorescence signals was normalized to make the initial value starts at zero. The detection time interval was 2â5âmin. The fluorescence results were obtained by averaging the values from three replicates of the experimental results.
Polyacrylamide gel electrophoresis experiments
The reactions of the methyl transfer were verified using native polyacrylamide gel electrophoresis (PAGE); 12% gels were prepared with 1à TAE/Mg2+ buffer with 12.5âmM MgCl2. All samples were run at 100âV for 1.5â2âh at 4â°C. After staining the polyacrylamide gels with Stain-All, the gels were imaged by a scanner (Canon LIDE 100). To detect FAM-modified DNA complexes, the gels were imaged directly under ultraviolet light without staining (Gel Image System Tanon-1600 or camera).
Agarose gel experiments
Asymmetric PCR41 experiments were verified by agarose electrophoresis; 1% agarose was prepared with 1Ã TAE buffer that had been supplemented with 2.5âmM MgCl2. All samples were run at 60âV for 1.5â2âh at room temperature. After staining the polyacrylamide gels with GelRed, the gels were imaged by Gel Image System Tanon-1600.
Preparation of ssDNA carriers
The designed ssDNA template carriers (T960 and T1200) were synthesized chemically and inserted into plasmid pUC57 (ref. 42). ssDNA templates were then prepared using an asymmetric PCR strategy, where the concentration ratio of forward to reverse primers is 60:1. The reactions were carried out in a PCR thermal cycler using the following protocol: 94â°C for 5âmin, then 30 cycles of 94â°C for 30âs, 60â°C for 30âs and 72â°C for 1âmin, thereafter 75â°C for 10âmin, and finally kept at 25â°C. To verify whether the purified gel band is the authentic target ssDNA carrier, we used FAM-modified complementary DNA probes to hybridize the target ssDNA. After adding fluorescent probes for 2âh at room temperature, target DNA complexes were separated by agarose electrophoresis (60âV for 1.5â2âh). After fluorophore labelling, target gel bands were collected by cutting the agarose gel (Extended Data Fig. 1).
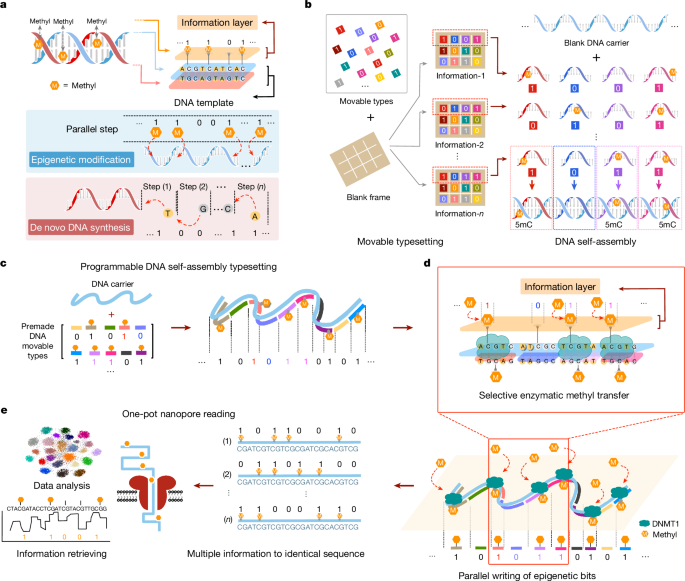
Programmable typesetting and methylation writing
The procedures of the standard methyl modification experiment were as follows: (1) a set of methylated and unmodified DNA brick strands was selected to mix with DNA carriers at 40:1 at a concentration of 0.4âμM in 1à TAE/Mg2+ buffer. Then, the sample was annealed in a PCR thermal cycler using the following protocol: 95â°C for 5âmin, 65â°C for 30âmin, 50â°C for 30âmin, 37â°C for 30âmin, 25â°C for 30âmin and finally kept at 25â°C. (2) Methyltransferase DNMT1 (refs. 43,44), SAM, methyltransferase reaction buffer and 50% glycerol were added to the reaction solution. The samples were then incubated at 37â°C for 3âh, 65â°C for 20âmin and finally kept at 25â°C. (3) The DNA complexes were gel purified for the next experiments.
GlaI-digestion-assisted detection of basic methylation writing
DNA complexes were produced by mixing DNA strands with equal molar concentrations (4âμM) in a 1à TAE/Mg2+ buffer. The annealing protocol was used for DNA complex assembly in fluorescence assay experiments. In fluorescence assay experiments, the reagents for basic methylation writing were mixed as follows: DNA complexes 1.5âμl, DNMT1 buffer 3âμl, SAM 3âμl, 50% glycerinum 3âμl, DNMT1 5âμl and H2O 14.5âμl. In PAGE experiments, the 30âμl basic methylation writing reaction included: DNA complexes 6âμl, DNMT1 buffer 3âμl, SAM 3âμl, 50% glycerinum 3âμl, DNMT1 5âμl and H2O 10âμl. After the basic methylation writing reactions, the mixture was incubated at 37â°C for 3âh, 3âμl GlaI buffer and the corresponding concentration of GlaI enzyme (5âUâμlâ1) were added21,45 (the volume of water was changed to keep the total volume constant at 30âμl). For fluorescent detection, the samples were placed in a fluorescence detector and reacted at 30â°C for 3âh. Fluorescence was detected every 3âmin. For PAGE analysis, 12% PAGE was used and the voltage was kept at 90âV for 70âmin.
High-throughput epi-bit DNA storage
The experimental procedure consisted of three parts: carrier T960 preparation, barcode connection and methyl writing. (1) Carrier preparation process: first, ssDNA carrier T960 was amplified from the plasmid by asymmetric PCR. Then the 5â² end of the carrier was phosphorylated by T4 polynucleotide kinase (PNK), followed by labelling of the target carrier T960 by fluorescent probes and purifying the carrier from agarose gels using the reagent kit (NucleoSpin Gel and PCR Clean-up, Mini kit). (2) Barcode connection: T960 carriers were specifically ligated with the 25 barcodes in individual tubes by adding T4 ligase at 25â°C for 30âmin, followed by inactivation at 65â°C for 20âmin. Then, 25 kinds of barcoded DNA carriers were purified by electrophoresis through a 1% agarose gel and recovery by the reagent kit. (3) Methyl writing: each of the 25 kinds of barcoded DNA carrier was mixed with a set of DNA bricks at a certain ratio and annealed. The methyl writing reaction was then carried out by adding DNMT1 methyltransferase at 37â°C for 3âh. Subsequently, the 25 kinds of carrier were mixed into one tube and purified using the reagent kit. Finally, the mixed sample of 25 kinds of carrier was prepared for nanopore sequencing.
Nanopore sequencing preparation
DNA samples were first prepared following the protocols of the Ligation Sequencing Kit (Oxford Nanopore Technologies (ONT), SQK-LSK109/SQK-LSK110) to construct the sequencing library, and then sequenced on MinION single-molecule sequencing device (ONT) by loading 50âfmol sample into a R9.4.1 flowcell. The device was operated using the bundled software MinKNOW to monitor running status. Base calling was done individually after sequencing using Guppy (ONT).
Methylation calling
Software Megalodon23 was used for methylation calling. Megalodon is an ONT-developed analysis tool that is capable of calling modified bases with high precision by anchoring the information-rich base-calling neural network output to a reference DNA sequence. Megalodon predicts methylation at both the per-read and per-site level (by aggregating per-read results) based on the log probability of whether or not the base is modified. The primary Megalodon run mode requires Guppy base caller (v.4.0 and above), and appropriate Rerio model is recommended for the accurate modified base calls46. In experiments, we used Megalodon v.2.5.0 with Guppy v.5.0.16 with a 5mC calling model (res_dna_r941_min_modbases_5mC_CpG_v001) from Rerio, and chose the default probability cutoff (0.8) to predict DNA methylation.
Coding strategy to store images of modified nucleotide structures
Chemical structure drawings of four DNA bases and their modified derivatives were first converted into 10âÃâ10 bitmap pictures, and then flattened to obtain binary sequences with a length of 100âbits. Next, a random seed was used to generate a sequence of binary numbers of equal length, which was then used to perform bitwise XOR operation with the sequence of information, to obtain a new sequence with changed distribution of 1âs. This process was performed iteratively until a sufficiently sparse sequence was obtained, namely, the proportion of epi-bit 1âs in the sequence was less than one-third and the maximum number of consecutive epi-bit 1âs is 3. Each of the sparse sequences was finally written into a group of 960 nt DNA carriers with the same loading sequence (Supplementary Fig. 16).
Information retrieval from high-throughput epi-bit storage
The sequence representing the original digital information was a stream of binary number, and bit-by-bit XOR was used to sparsify the sequence. Therefore, each methylation site corresponded to one pixel in the original binary images. This correspondence produced a grey-scale image as the methylation predictions obtained from Megalodon are probabilities (0â100%). Restoring these images required de-sparsification. Specifically, for each bit (pixel), if 0 is used for performing XOR for an epi-bit site, the pixel value in grey-scale image is exactly the methylation probability. In the opposite circumstances, the value in grey-scale image is 1 minus the methylation probability of the corresponding methylation site (Supplementary Fig. 29). After these conversions, grey-scale images were binarized to bitmaps by setting an appropriate threshold.
Determination of the threshold of epi-bit calling
To determine the optimal threshold for calling epi-bits 1 and 0 from methylation probability data, we conducted analysis for the 32 methylation sites on the T960 DNA carriers individually. Specifically, for each methylation site, we assumed that both the probability values detected in nanopore sequencing of the status 0 (no methylation) and the status 1 (methylation) follow the Gaussian distribution. Therefore, the probability distribution detected from a methylation site was supposed to be the sum of two independent Gaussian distributions,
$${{\rm{B}}{\rm{G}}}_{{i}}(x,\alpha ,{\mu }_{i1},{\mu }_{i2},{\sigma }_{i1},{\sigma }_{i2})={\alpha }_{i}\frac{1}{{\sigma }_{i1}\sqrt{2\pi }}{e}^{-\frac{{(x-{\mu }_{i1})}^{2}}{{2{\sigma }_{i1}}^{2}}}+(1-{\alpha }_{i})\frac{1}{{\sigma }_{i2}\sqrt{2\pi }}{e}^{-\frac{{(x-{\mu }_{i2})}^{2}}{{2{\sigma }_{i2}}^{2}}}$$
where for each methylation site i, μi1 and μi2 are the mean and s.d. of the two Gaussian distributions for status 0 and status 1, respectively, and Ïi1 and Ïi2 are the s.d. of the two Gaussian distributions for status 0 and status 1, respectively. αi is the fraction of status 0 in all epi-bits printed at site i. To obtain the values of the above parameters, we fitted the above functions with the probability distribution obtained from each methylation site. It is worth noting that μi1 should be close to 0 (as the unmethylated position is unlikely to generate a higher methylation signal), and μi2 should be a higher value, otherwise the fitting will generate only a single peak. After fitting, each of the 32 methylation sites generated the corresponding double peak distribution (Extended Data Fig. 5). Next, we defined an objective function on threshold,
$${\rm{OBJ}}({\rm{th}})=-\mathop{\sum }\limits_{i=1}^{32}\left({\int }_{0}^{{\rm{th}}}\frac{1}{{\sigma }_{i1}\sqrt{2\pi }}{e}^{-\frac{{(x-{\mu }_{i1})}^{2}}{{2{\sigma }_{i1}}^{2}}}+{\int }_{{\rm{th}}}^{1}\frac{1}{{\sigma }_{i2}\sqrt{2\pi }}{e}^{-\frac{{(x-{\mu }_{i2})}^{2}}{{2{\sigma }_{i2}}^{2}}}\right)$$
Then, we maximize the objective function to obtain the optimal threshold.
Automatic sampling for data writing
Data writing of the pictures of the tiger rubbing and the panda were performed on an automated four-channel liquid handling system by HCSCI China. Briefly, the stock solution of 175âÃâ4â=â700 bricks was loaded into eight 96-well plates (source plates). Specific combinations of stock solutions were added to destination wells on 384-well plates, where each well held all bricks necessary to guide data writing on five templates L1âL5. A sampling sheet was prepared based on the epi-bit information of the data to be stored. The sampling sheet dictated the sampling scheme from the source wells to the destination wells. The liquid handling system then dispenses 500ânl of each source solution to each destination well according to the sampling sheet. Each destination well held 175âÃâ0.5â=â87.5âμl of mixture brick solution after the sampling was completed. The carriers, enzymes and reaction buffers were then added to all destination wells for data writing.
Coding strategies for data compression and error correction for the tiger rubbing and panda
The original image was first read as a binary stream. This binary stream was compressed. Next, BCH code was used to add logic redundancy to the compressed information. Specifically, the information was divided into groups. Each of these groups was used as information symbols to generate redundancy, which resulted in a coding matrix. Next, this matrix was transposed and flattened, resulted in a binary stream.
All the barcodes used for storing information were 20âbits in length. A seed barcode was first generated (for example, 01110101001011001001), and then barcodes with random bits were generated. Next, each barcode was verified to conform to the following rule. The valid barcode was recorded only when the minimum Hamming distance between it and all the recorded barcodes is greater than four. A total of 370 valid barcodes were selected, of which 250 barcodes possessed a 1 ratio of 40â60%. After site optimization (Extended Data Fig. 7), 16 sites were dropped and 5âÃâ20â=â100 sites were selected as barcode sites; thus, there were 234 sites for storing the image data. The compressed binary stream was divided into groups (234âbits per group). Depending on the barcode generation strategy, barcodes were selected randomly and assigned to groups. Finally, all groups were stored in wells.
The retrieved binary stream was first truncated, and then rearranged to a matrix. Next, this matrix was transposed, and each row of this matrix was used as a decoding unit. After BCH decoding, this matrix was flattened, resulted in a binary stream. Finally, the binary stream was visualized as the stored image.
Simulations of error correction capabilities in large-scale epi-bit DNA storage
In silico simulation was performed to test the capacity of error correction for epi-bit DNA storage, where the epi-bit information of tiger rubbing (Fig. 5e) and panda (Supplementary Fig. 42) were simulated independently. For all simulation, assuming that 50âbits of information was loaded on each DNA carrier, and errors were distributed independently on different DNA carriers. The error frequency was sampled from a pre-experiment, in which 48 wells (240 DNA carriers) were sequenced collectively with nanopore sequencing. The fluctuations of error rate were realized by adding or deleting single epi-bit error manually on DNA carriers.