Mice
All animal care and experimental procedures were ethically performed and approved by the Institutional Animal Care and Use Committee at Rockefeller University. Male mice were single-housed with a 12âh light/12âh dark cycle and ad libitum access to regular chow and water, except in fasting and DIO studies, where either a HFD with 45âkcal%fat (4.7âkcalâgâ1) or a HPD with 42âkcal%fat and high sucrose content (4.5âkcalâgâ1) (TD.88137, Envigo) was provided. We used male ob/ob (B6.Cg-Lepob/J; 000632, Jackson Laboratory; or bred in-house), Rosa26fsTRAP (B6.129S4-Gt(ROSA)26Sortm1(CAG-EGFP/Rpl10a,-birA)Wtp/J; 022367, Jackson Laboratory), AgRPâCre (AgRPtm1(cre)Lowl/J; 012899, Jackson Laboratory) and Flp reporter mice (RCF-tdTomato, B6.Cg-Gt(ROSA)26Sortm65.2(CAG-tdTomato)Hze/J; 032864, Jackson Laboratory) crossed to Cre reporter mice (Rosa26fsTRAP). BDNFâIRESâCre mice57 were provided by W. Shen (Shanghai Institute of Technology). POMC Dre mice were provided by J. Bruning (Max Planck Institute for Metabolism Research). For retrograde tracing from motor neurons in Mo5, ChatâCre mice (B6.129S-Chattm1(cre)Lowl/MwarJ; 031661, Jackson Laboratory) were crossed to Helper RabV mice (B6;129P2-Gt(ROSA)26Sortm1(CAG-RABVgp4,-TVA)Arenk/J; 024708, Jackson Laboratory) and TrkBF616A knock-in mice carrying a point mutation that renders the receptor sensitive to an allele-specific kinase inhibitor (1-NM PP)58. All mouse lines were in a WT (C57BL/6âJ) background. For brain surgeries, male mice of at least 8 weeks of age were anesthetized with isoflurane and placed in a stereotaxic frame (David Kopf Instruments), a craniotomy was performed, and a borosilicate glass pipette was used to inject viral vectors. For VMH injections: three injections (each 50ânl) were made into each hemisphere (bregma, â1.36âmm; midline, ±0.35âmm; from brain surface, 5.70âmm, 5.60âmm and 5.50âmm). For injections into the Arc, 50ânl was injected as follows: bregma, â1.45âmm; midline, ±0.45âmm; from brain surface, 5.70âmm, 5.60âmm and 5.50âmm. For injections into Mo5, 75ânl was injected as follows: bregma, â5.20âmm, midline, ±1.5âmm; from brain surface, 4.60âmm and 4.50âmm. For Me5 injections: from bregma, â5.4âmm; midline, ±0.9âmm; from brain surface, 4.5âmm, 4.0âmm, 3.5âmm.
Reagents
Leptin was diluted in sterile saline (3âmgâkgâ1) and injected intraperitoneally. For photometry recording, mice were fasted overnight and injected with either saline or leptin 2âh before recordings. All mice received saline and leptin injections in alternating order. For acute food intake experiments, leptin or saline was injected 2âh before onset of the dark period, and all mice received saline and leptin injections in a cross-over design.
Viruses
Cre-dependent neuronal ablation was performed by injection of AAV1-mCherry-flex-dtA (UNC Vector Core)59. To target expression of calcium activity indicator GCaMP6s to VMHBDNF neurons, we used an AAV vector carrying a double-floxed GCaMP6s construct (AAV5-Syn-Flex-GCaMP6s-WPRE-SV40, Addgene)60. For optogenetic manipulations, a somatic targeting GtACR (AAV5-hSyn1-SIO-stGtACR1-FusionRed)61 or ChR (AAV5-EF1a-double-floxed-hChR2(H134R)-EYFP-WPRE-HGHpA)62 was used (both Addgene). For long-term silencing, a Cre-dependent TelC AAV (AAV5-hSyn-FLEX-TeLC-P2A-dTomato, Addgene) was used, and for long-term activation a Cre-dependent NaChBac (AAV-Syn-DIO-NaChBac-dTomato) and a Dre-dependent NaChBac63 (AAV5-hSyn-roxSTOProx-NaChBac-dTomato, HHMI-Janelia Research Campus) were used. For retrograde tracing, a combination of two helper AAVs (AAV1-TREtight-mTagBFP2-B19G and AAV1-syn-FLEX-splitTVA-EGFP-tTA, both Addgene) and pseudotyped Rabies (EnvA G-Deleted Rabies-mCherry, Salk Institute)64,65 were injected; for retrograde labelling from Mo5, a G-deleted Rabies-H2B-mCherry (Salk viral core) was used; and for anatomical tracing from Me5, a retrograde mCherry construct (pAAV-hSyn-DIO-hM4D(Gi)-mCherry, Addgene) was used. For projection activation, we used an AAV encoding eOPN3 (AAV-hSyn1-SIO-eOPN3-mScarlet-WPRE, Addgene)61, and for labelling projections with ChR we used a retro-AAV (AAV-EF1a-double-floxed-hChR2(H134R)-mCherry-WPRE-HGHpA). Anterograde labelling was done with AAV1 (ref. 66) encoding Cre (AAV-hSyn-Cre-WPRE-hGH, Addgene) and Flp (AAV-EF1a-Flpo, Addgene). For âCre-outâ experiments, AAV-Ef1a-DO-ChETA-EYFP-WPRE-pA (Addgene) was used.
Immunofluorescence
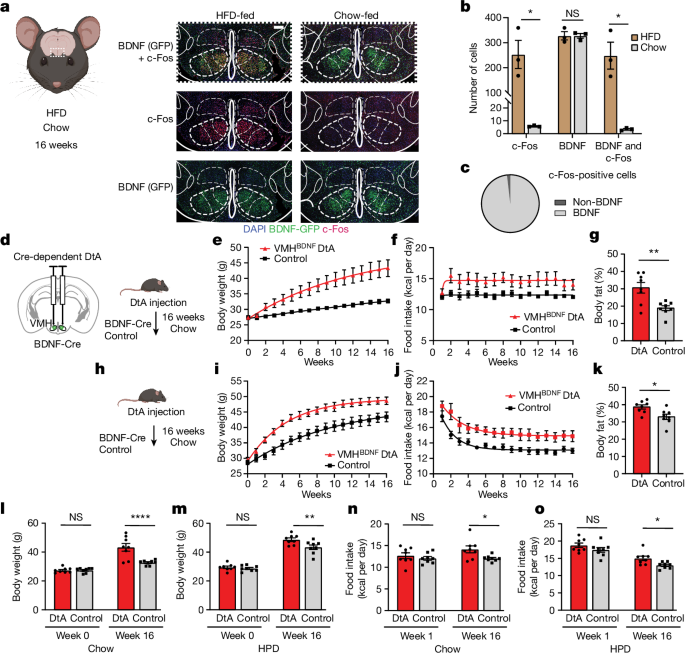
For c-Fos staining after DIO, BDNFâCre mice were crossed to Rosa26fsTRAP to express eGFP in a Cre-dependent manner in BDNF neurons. Mice were fed a HFD, while littermate control mice were fed chow. After 16 weeks, mice were transcardially perfused with 4% paraformaldehyde, and their brains were postfixed for 1 day in 4% paraformaldehyde. Brains were then placed in 30% sucrose in phosphate-buffered saline (PBS) until precipitation and frozen and coated in OCT for cryosectioning. Cryosections (50âμm) were cut using a Leica cryostat (CM1950). Brain sections were washed in PBS with 0.1% Triton X-100 (PBST, pH 7.4) and blocked in 3% normal goat/donkey serum (Jackson ImmunoResearch Laboratories) and 2% BSA (Sigma) in PBST for 2âh. Slides were then incubated overnight at room temperature with primary antibody. After being washed in PBST, sections were incubated with fluorescein-conjugated goat IgG. The primary antibodies used and their dilutions were as follows: rabbit anti-FOS (1:1,000; mAb 2250S, Cell Signaling), chicken anti-GFP (1:1,000, ab13970, Abcam). Secondary antibodies conjugated with Alexa-594 and Alexa-488 were purchased from Invitrogen. Brain sections were mounted on to SuperFrost (Fisher Scientific 22-034- 980) slides and then visualized with an inverted Zeiss LSM 780 laser scanning confocal microscope with a Ã10 or Ã20 lens. Images were imported to Fiji for further analysis and to count cells. To quantify numbers of stained cells, brain slides were imaged under a Ã20 objective. For advillin staining, the procedure was the same as above but with rabbit anti-advillin (1:500, NBP2-92263, Novus Biologicals) as the primary antibody and Alexa 647 donkey anti-rabbit (1:500, ab150075, Abcam) as the secondary antibody. For anterograde tracing, brains were processed as described above; tdtomato/Ruby and GFP were amplified with rabbit anti-RFP (1:1000, 600-401-379, Rockland) and chicken anti-GFP (1:1,000, ab13970, Abcam) as primary antibodies, and secondary antibodies conjugated with Alexa-594 and Alexa-488 were purchased from Invitrogen.
In situ hybridization
Mice were briefly transcardially perfused with RNase-free PBS to remove blood. Brains were then quickly collected, frozen in OCT and stored at â80â°C until they were sectioned by cryostat (15âμm sections) and attached on Superfrost Plus Adhesion Slides (Thermo Fisher). RNAscope Fluorescent Multiplex assay (Advanced Cell Diagnostics Bio) was then performed using the RNAscope system as per the manufacturerâs protocol. Probes for the following mRNAs were used (all from ACDBio): mm-BDNF (catalogue no. 424821) and eGFP (catalogue no. 400281), VGlut2 (catalogue no. 319171), RabV (catalogue no. 456781), AgRP (catalogue no. 400711), POMC (catalogue no. 314081), MC4R (catalogue no. 319181-C2) and NPY5R (catalogue no. 589811), LepR (catalogue no. 402731). Briefly, a hydrophobic barrier was created using Immedge Hydrophobic Barrier Pen (Vector Laboratories). Slides were pretreated by serial submersion in 1à PBS, 50% EtOH, 70% EtOH and twice 100% EtOH for 2âmin each, at room temperature. Probe hybridization was achieved by incubation of 35âμl mRNA target probes for 2âh at 40â°C using a HyBez oven. The signal was amplified by subsequent incubation of Amp-1, Amp-2, Amp-3 and Amp-4, one drop each, for 30, 15, 30 and 15âmin, respectively, at 40â°C using a HyBez oven. Each incubation step was followed by two 2âmin washes with RNAscope washing buffer. Nucleic acids were stained using DAPI Fluoromount-G (SouthernBiotech) mounting medium before coverslipping. Slides were visualized with an inverted Zeiss LSM 780 laser scanning confocal microscope using a Ã20 or Ã40 lens. Images were imported to Fiji for further analysis.
Long-term body weight and food intake measures
Single-housed mice were measured weekly to assess body weight and food intake. Whole-body composition was measured using nuclear magnetic resonance relaxometry (EchoMRI) at the end of the 16 week period.
Optogenetics
After injection of AAVs encoding either ChR or GtACR, we bilaterally implanted 200âμm fibre optic cannulas (Thorlabs) in BDNFâCre mice and control mice (Cre-negative littermates). For VMH targeting, implants were angled at 15° and placed at the following positions: bregma, â1.36âmm; midline, ±1.85âmm; from brain surface: 5.25âmm. For brainstem targeting, implants were angled at 15° and placed at the following positions: bregma, â5.4âmm; midline, ±1.75âmm; from brain surface, 2.6âmm (for Me5); bregma, â6.3âmm; midline, ±2.15âmm; from brain surface, 5.5âmm (for LPGi); and bregma, â5.7âmm; midline, ±2.6âmm; from brain surface, 4.7âmm (for PCRT). Implants were subsequently fixed with dental cement (C&B Metabond). After a minimum of 3 weeks expression time, mice were handled and habituated to tethering with optical fibres. A constant 473ânm laser (OEM Lasers/OptoEngine) was used for optogenetic inhibition with GtACR and pulsed at 2âHz (5âms) for optoactivation with ChR. For inhibition with OPN, a 532ânm laser at 10âHz was used. Lasers were connected to bifurcated optical fibres (Thorlabs) with an output of approximately 2â5âmW at the implant. For AgRP projection stimulation, the laser power was reduced to 1â2âmW. Food intake studies were done in home cage-like arenas during the light phase without bedding unless otherwise stated.
Acute food intake experiments
With optogenetic activation or inhibition, mice were habituated to the arena for 10âmin without food present. Then, consumption of a single food pellet was measured every 30âmin for five repetitions, with only the second repetition being paired with optogenetic activation or inhibition. For open loop and closed loop feeding experiments, a single chow pellet was fixed to the middle of a home cage-style arena with fun-tak (Loctite). Food intake was assessed every 5âmin in three repetitions, with only the second repetition being paired with optogenetic inhibition. Inhibition was either 5âmin constant laser (open loop) or triggered (closed loop) by real-time video tracking (Noldus, Ethovision) whenever the head of the mouse was within a 3âcm radius of the pellet. For modification with bedding present, the same open loop set-up was used but with corn cob bedding covering the floor. For wood block trials, the chow pellet was replaced by a wood block that was fixed with fun-tak. Time spent biting the wood block was manually assessed and quantified by scoring of video recordings.
Liquid diet experiments
Ensure Vanilla (20âµl) was pipetted on to the bare floor of a cage in three repetitions without light activation, followed by three repetitions with light activation and another three repetitions without light activation. For quantification purposes, experiments were video recorded, and latencies from Ensure delivery to full consumption were scored and averaged over the three repetitions.
Operant conditioning
Trials were performed in a home cage-style arena with two capacitive touch plates mounted on opposite sides. Both touch plates were connected to an Arduino to register numbers of touches, and one randomly assigned side would trigger cessation of a constant 2âHz laser (for ChR) for 3âs or activation of a constant laser for 3âs (for GtACR). Trials lasted for 1âh.
Conditioned flavour preference assays
Assays were performed as previously described44. Briefly, mice were habituated overnight to orange- and strawberry-flavoured sugar-free Juicy Gels (Huntâs). Initial preference was assessed in a 30âmin session without any light application. The preferred flavour was then paired with light exposure for ChR mice, or the less preferred flavour paired with light exposure for GtACR mice. Conditioning was repeated daily for 3 days and consisted of one light exposure session in which light exposure started after 5âmin and lasted for 25âmin while the paired gel was presented and a 30âmin session with the non-paired gel without any light exposure. A 15âmin test session in which both gels were available was performed on the day after conditioning ended.
Spaghetti and wooden stick experiments
Five spaghetti sticks or wooden sticks of similar length were distributed equally in an empty home cage. Control and eOPN3 mice were given 5âmin baseline exploration time, 5âmin with laser inhibition and 5âmin without laser with the spaghetti or sticks present. A side and overhead camera were used to quantify the time spent chewing.
Head-fixed jaw-tracking
Mice for head-fixed experiments had a small metal bar fixed to their skull with dental cements during implant surgery. After a minimum of 3 weeks recovery, mice were habituated to being head-fixed in a custom head-fixation set-up. This set-up consisted of a side camera (Basler a2A1920-160umPRO -ace 2) and a laser source controlled and synchronized by Bonsai67. Frames were acquired at 100âHz at 722âÃâ878 pixel size. Optogenetic inhibition trials consisted of 1âmin with laser, 1âmin on and 1âmin off. Jaw pose was subsequently estimated with DeepLabCut68.
Fibre photometry
After injection of an AAV encoding Cre-dependent GCaMP6s into the VMH of male BDNFâCre mice, a unilateral 400âμm fibre optic cannula was implanted as described for optogenetics. After a minimum of 4 weeks expression time, mice were habituated to tethering and a home cage-style arena.
Data were collected with a Fiber Photometry system by Tucker-Davis Technologies (RZ5P, Synapse), and Doric components and recordings were synched to video recordings in Ethovision by TTL triggering. A 465ânm and isosbestic 405ânm LED (Doric) were reflected into a dual fluorescence Mini Cube (Doric) before entering the recording fibre that connects to the implant. Recording fibres were photobleached overnight before recordings to minimize autofluorescence. GCaMP6s fluorescence was collected as a calcium-dependent signal (525ânm) and isosbestic control (430ânm) with a femtowatt photoreceiver (Newport, 2151) and a lock-in amplifier using the RZ5P at a 1âkHz sampling rate.
Mice were allowed to habituate for 30âmin at the start of each recording session before any items were introduced into the arena. Feeding bouts were manually assessed and scored from video recordings when mice were given single pellets of chow or 20âmg sucrose treat pellets (Bio-Serv). Instances of food interaction without consumption were defined as approach within a 2âcm radius around a chow pellet without subsequent consumption. To measure the effects of different energy states, the same mice underwent the same standardized recording procedure in the following states: lean ad lib chow-fed, overnight fasted injected with saline, and overnight fasted injected with leptin and 4 weeks DIO. The order of the lean, fasted saline and fasted leptin states was randomized to avoid any order effects.
A script written in MATLAB based on a previously published method and code was used for analysis69. Bleaching and movement artefacts were removed by applying a polynomial least-squares fit to the 405ânm signal, adjusting it to the 465ânm trace (405fitted) to then calculate the GCaMP signal as %ÎF/Fâ=â(465signalâââ405fitted)/405fitted. Traces were filtered with a moving average filter and downsampled by a factor of 20. Three trials per mouse were averaged to derive data for peri-event plots and analysis of maximum and minimum signals.
Quantification and statistics
Sample sizes were chosen on the basis of similar studies previously published and kept to a minimum to reduce unnecessary use of animals. Experimenters were blinded to group allocation as much as possible, but small groups sizes and concurrent recordings of control and treatment animals meant it was sometimes not possible. Group allocation was done at random, unless genetic backgrounds dictated group assignment. Microscopy images were analysed and quantified in ImageJ/Fiji. Photometry recordings were processed and analysed with MATLAB (MathWorks). Statistical analyses were performed in GraphPad Prism. All tests were two-sided, and results are displayed as meanâ±âs.e.m. Statistical details are provided in the figure legends and source data, including definitions of n and significance. Significance was defined as Pâ<â0.05. Mice were randomized into control or treatment groups. Control mice were age-matched littermate controls where possible. Graphs were produced using GraphPad Prism and Adobe Illustrator, and schematic illustrations were prepared in BioRender.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.