Tissue acquisition and processing
Human developmental tissue samples used for this study were obtained from the MRCâ Wellcome Trust-funded Human Developmental Biology Resource (http://www.hdbr.org) with approval from the Newcastle and North Tyneside NHS Health Authority Joint Ethics Committee (08/H0906/21+5) and East of EnglandâCambridge Central Research Ethics Committee (NHS REC 96/085). Prenatal skin for immunofluorescence imaging of proteins causing congenital skin disorders were obtained with approval from the Guyâs and St Thomasâ Hospital Trust Ethics Committee. For samples used for 3D rendering in the Koehler Laboratory, fetal tissue specimens were obtained from the Birth Defects Research Laboratory at the University of Washington (UW) with approval from the UW institutional review board Committee, and the study was performed in accordance with ethical and legal guidelines of the Boston Childrenâs Hospital institutional review board. All samples were collected following either elective termination of pregnancy or miscarriages, with informed written consent, following all relevant rules and regulations.
Tissues were processed into single-cell suspensions immediately after receipt for single-cell transcriptomic profiling. Tissue was first transferred to a sterile 10âmm2 tissue culture dish and cut in <1âmm3 segments using a scalpel. It was then digested with typeâIV collagenase (final concentration of 1.6âmgâmlâ1; Worthington) in RPMI (Sigma-Aldrich) supplemented with 10% heat-inactivated FBS (Gibco) at 37â°C for 30 min with intermittent agitation. Digested tissue was then passed through a 100âμm cell strainer. For 2 samples (F220, F221), 500âµl of 0.25% trypsin (Sigma-Aldrich) was further added to any remaining unfiltered tissue and incubated at room temperature for 5âmin. Cells were collected by centrifugation (500g for 5âmin at 4â°C). Cells were treated with 1à RBC lysis buffer (eBioscience) for 5âmin at room temperature and washed once with flow buffer (PBS containing 5% (v/v) FBS and 2âmM EDTA) before cell counting and antibody staining. Single-cell suspensions were generated from skin of 18 donors with ages spanning from 7âPCW to 17âPCW.
scRNA-seq experiment
Dissociated cells were stained with anti-CD45 antibody (1:20, PE, clone HI30, BD Biosciences (samples F220 and F221) or 1:33, BUV395, clone HI30, BD Biosciences (other sorted samples)) on ice in the dark for 30âmin, except for one sample (F217) for which no cell sorting was performed. To improve capture of less abundant cell populations from the CD45â fraction, such as keratinocytes and endothelial cells, additional staining was carried out to separate them from the abundant CD34+ stromal cells in a subset of samples. For samples F220 and F221, anti-CD34 (1:25, APC/Cy7, clone 581, BioLegend) antibodies was used; for samples F69 and F71, additional staining included anti-CD34 (1:25, APC/Cy7, clone: 581, BioLegend) and anti-CD14 antibodies (1:33, PE-CF594, clone MÏP9, BD Biosciences). Immediately before sorting, cells were passed through a 35âµm filter (Falcon), and DAPI (Sigma-Aldrich) was added at a final concentration of 3âμM. Sorting by flow cytometry was performed with a BD FACSAria Fusion flow cytometer. The CD45+ fraction was sorted from the DAPIâCD45+ gate, and the CD45â fraction was sorted from the DAPIâCD45â gate. CD45 gating was contiguous so that no live cells were lost in sorting. Live CD45+ and CD45â cells were sorted into separate chilled FACS tubes coated with FBS. For samples F220 and F221, CD34+ and CD34â fractions were sorted from CD45âCD34+ and CD45âCD34â gates, respectively. For samples F69 and F71, in addition to the live CD45+ and CD45â cells, we isolated all cells from the CD45â fraction that were not within the CD34+CD14â gate and collected them into a separate chilled FACS tubes coated with FBS (Extended Data Fig. 1a).
FACS sorted cell suspensions were counted and loaded onto a 10x Genomics Chromium Controller to achieve a maximum yield of 10,000 cells per reaction. Either Chromium single-cell 3â² reagent kits (v.2) or Chromium single-cell V(D)J kits from 10x Genomics were used. Cells were loaded onto each channel of the Chromium chip following the manufacturerâs instructions before droplet encapsulation on the Chromium Controller. Gene expression and TCR libraries were generated according to the manufacturerâs instructions. The gene expression libraries were sequenced to achieve a minimum target depth of 20,000 reads per cell and the TCR libraries were sequenced to achieve a minimum target depth of 5,000 reads per cell using Illumina sequencing instruments.
Statistics and reproducibility
Images of haematoxylin and eosin-stained skin sections (Fig. 2a) were taken from 13 independent samples from the following gestational ages: 6âPCW (nâ=â1 and 3 sections), 8âPCW (nâ=â3), 11âPCW (nâ=â2), 14âPCW (nâ=â1 and 2 sections), 15âPCW (nâ=â4) and 17âPCW (nâ=â2).
Image analysis of multiplex RNAscope and immunofluorescence staining was performed on independent biological and/or technical replicates for each experiment: nâ=â5 biological replicates for RNAscope slides with FOXP3, SHH, SLC26A7 and NDP probes (Fig. 2c); nâ=â1 biological replicate with 4 technical replicates for RNAscope slides with ACKR3, CXCL12, PDGFD and SERPINB7 probes (Fig. 2h); nâ=â3 biological replicates with 2 technical replicates for immunofluorescence slides with anti-FOXP3, anti-SOX2 and anti-KRT14 (Extended Data Fig. 3h); nâ=â1 biological replicate with nâ=â2 technical replicates for immunofluorescence slides with anti-LYVE1, anti-CD45 and anti-VIM (Fig. 3e); nâ=â3 biological replicates for RNAscope slides with CDH5, CD68, P2RY12 and ELAVL3 probes (Fig. 4a); nâ=â1 biological replicate with 4 technical replicates for immunofluorescence slides with anti-CD45, anti-LYVE1 and anti-CD31 (Fig. 4a); nâ=â15 biological replicates with a minimum of nâ=â2 technical replicates for prenatal skin immunofluorescence slides with each of the following antibodies: anti-KRT1, anti-KRT14, anti-plectin, anti-BP180, anti-laminin-332 and anti-typeâVII collagen (Extended Data Fig. 6e); nâ=â3 biological replicates for prenatal skin whole-mount immunofluorescence with anti-CD31 and anti-LYVE1 (Fig. 4a); whole-mount immunostaining of SkO co-culture with and without macrophages was performed on nâ=â5 SkOs without macrophages and nâ=â5 SkOs with macrophages (Fig. 4f); immunostaining of cryosections of SkOs co-cultured with macrophages was performed on nâ=â2 SkOs (Fig. 4h).
Visium spatial transcriptomic data were generated from nâ=â4 biological samples from 3 different sites, with 2 or 3 technical replicates each (Figs. 2f and 3d).
Scratch wound assays were performed on SkO-derived fibroblasts: nâ=â3 and each experiment included technical replicates in 3â6 wells (Extended Data Fig. 8g). Data represented as the meanâ±âs.d. and statistics generated with two-way analysis of variance (ANOVA) with Tukeyâs multiple comparisons test. Endothelial cells and macrophages were generated across two independent differentiation batches. Endothelial cell and macrophage co-culture for angiogenesis assays were performed on nâ=â6 wells (Extended Data Fig. 11i). Data represented as the meanâ±âs.d. and statistics generated with an unpaired t-test.
Human iPS and ES cell line information
The iPS cell line Kolf2.1S was obtained from the HipSci Initiative under a material transfer agreement. This line was not independently authenticated. Details about the generation and characterization of the line at the time of derivation is available from the HipSci website (https://www.hipsci.org/#/lines/HPSI0114i-kolf_2).
The WTC-mEGFP-DSP-cl65 iPS cell line and the WA25 ES cell lines were obtained under a material transfer agreement with the Coriell or WiCell Institute. These lines were determined to have a normal karyotype before SkO differentiation.
All cell lines tested negative for mycoplasma before experiments.
Scratch wound assay of fibroblasts in co-culture with macrophages
Fibroblasts were isolated from Kolf2.1S-derived SkOs (nâ=â10) at dayâ76. In brief, SkOs were washed with dPBS then incubated with dispase and a ROCK inhibitor for 40âmin at 37â°C. The epidermis and dermis layers of the SkO were separated using forceps, and the dermis was transferred to collagenase for 40âmin at 37â°C. Collagenase was neutralized with fibroblast medium, and the single-cell suspension was filtered through a 40âµm cell strainer. After centrifugation at 180g for 3âmin, the fibroblasts were resuspended and seeded in fibroblast medium then cultivated as primary fibroblasts. Macrophages were differentiated from Kolf2.1S iPS cells as previously described80.
For the scratch assay, fibroblasts and macrophages were seeded in 48-well plates at 5:1 ratio then incubated for 24âh at 37â°C. The next day, the scratch was generated using a p1000 tip down the centre of each well. The assay was imaged using Incucyte S3, whole well module and analysed using the ImageJ Wound_healing_size_tool_updated macro81. Two-way ANOVA was carried out to assess statistics in GraphPad. Scratch assays performed were nâ=â3 independent experiments with 3â6 replicates per experiment.
Visium spatial data generation
Prenatal facial (nâ=â1, replicateâ=â2) and abdominal skin (nâ=â1, replicateâ=â2) samples from a single donor at 10âPCW were embedded in optimal cutting temperature (OCT) medium and flash-frozen in isopentane cooled with dry ice. Cryosections (10âµm) from the OCT blocks were cut onto 10x Genomics Visium slides. Sections were stained with haematoxylin and eosin and imaged at Ã20 magnification on a Hamamatsu Nanozoomer. These sections were then processed according to the 10x Genomics Visium protocol, using a permeabilization time of 12âmin found through a previous tissue optimization step. Dual-indexed libraries were prepared as per the manufacturerâs protocol, pooled at 2.8ânM and sequenced in 8 samples per Illumina Novaseq S4 flow cell with the following run parameters: readâ1: 28 cycles; i7 index: 10 cycles; i5 index: 10 cycles; read 2: 90 cycles.
Endothelial cell and SkO co-culture with macrophages
Endothelial cell culture co-culture and image acquisition
Endothelial cells were derived from Kolf2.1S iPS cells cultured on Matrigel-coated plates in mTeSR1 medium with ROCK inhibitor at 4.5âÃâ104 cells perâcm2. iPS cells were differentiated through lateral mesoderm into CD144+ endothelial cells as previously described82. Macrophages and SkOs were also derived from Kolf2.1S iPS cells according to previously published methods1,80. The angiogenesis assay was carried out by culturing iPS cell-derived endothelial cells and macrophages separately or in co-culture in 15-well 3D chambered µ-slide (ibidi, 81506). This was done using a three-layered sandwich method, whereby layer one was 10âµl Matrigel (Corning, 354230), layer two was supplemented StemPro medium (Gibco, 10639011)â+â10% Matrigel with and without the endothelial cells and layer three was supplemented StemPro medium with and without macrophages. The endothelial cells were left to settle for 4âh at 37â°C before addition of macrophages. The assay was imaged 2âh after initial culture and then every 24âh for 3âdays using an EVOS 7000 microscope, and images were analysed using Fiji distribution of the ImageJ software (v.2.14.0)83. Before co-culture, iPS cell-derived macrophages were phenotyped using flow cytometry (Extended Data Fig. 11h). Macrophages were collected using TrypLE (Gibco) at 37â°C, 5% CO2 for 5âmin, and cells were collected by centrifugation (300g for 6âmin). Cells were washed once with cell staining buffer (BioLegend) before cell counting and antibody staining. Nonspecific bindings were blocked using Human TruStain FcX (Fc receptor blocking solution, BioLegend) for 10âmin on ice and then stained using a Fixable Blue Dead Cell Stain kit for 10âmin on ice (1:500 in PBS, ThermoFisher). Cells were washed twice with cell staining buffer. Single-staining was performed on cells with anti-CD206 antibody (1:200, PE, clone 19.2, ThermoFisher), anti-CD16 antibody (1:50, PE-Cyanine7, clone eBioCB16, ThermoFisher), anti-CD14 antibody (1:100, PerCP-Cyanine5.5, clone 61D3, ThermoFisher), anti-CD1c antibody (1:25, Pacific Blue, clone L161, BioLegend), anti-CD45 (1:300, BV480, clone HI30, BD Biosciences) and anti-human Lineage Cocktail (1:100, CD3, CD19, CD20, CD56, clones UCTH1, HIB19, 2H7, 5.1H11, BioLegend) on ice in the dark for 30âmin. Before acquiring on the analyzer, cells were washed once in cell strained buffer and passed through a 35âµm filter (Falcon). Acquisition by flow cytometry was performed using a Cytek Aurora. Live single CD16+, CD14+, CD206+, CD45+, CD1câ and Linâ cells were analysed using FlowJo (v.10.9.0).
SkO co-culture and image acquisition
The co-culture was performed by adding the macrophages to the SkOs on dayâ12 of culture, with a 1:5 ratio. SkOs were transferred to a low attachment 96-well plate (Nunclon Sphera, Life Technologies) in SkO maturation medium1 containing 20% Matrigel (Corning). Macrophages were added to the SkOs and the co-culture was centrifuged at 100g for 6âmin 1âacc, 0âdec. On dayâ3 of co-culture, the cells were transferred to a low-attachment 24-well plate, and Matrigel was diluted with fresh SkO maturation medium. On dayâ35 of co-culture (dayâ47 of SkO differentiation), the SkOs were fixed in a 2âml tube with 4% paraformaldehyde (PFA) overnight for whole-mount serial staining (2 batches of differentiation, SkOs with macrophages nâ=â5, SkOs without macrophages nâ=â5). The co-culture was then permeabilized in blocking buffer (0.3% (v/v) Triton X-100, 1% (v/v) normal goat serum based on the antibodies and 1% BSA (v/v) dissolved in 1à PBS) for 8âh at room temperature on a shaker. Cells were then incubated overnight at 4â°C on a shaker (65âr.p.m.) with the first primary antibody, anti-CD45 (1:100, clone YAML501.4, ThermoFisher) for macrophages, for 48âh. The morning after, cells were washed and then incubated with the first secondary antibody overnight (goat anti-rat IgG, Alexa Fluor Plus 647, ThermoFisher). The morning after, SkOs were washed and incubated with the second primary antibody, anti-CD31 (1:100, clone JC70A, Dako) for endothelial cells for 48âh. The SkOs were then washed and incubated with the second secondary antibody (goat anti-mouse IgG1, Alexa Fluor 568, ThermoFisher) and DAPI overnight on a shaker. Cells were washed and placed in 50% glycerol for 30âmin on a shaker at room temperature. Cells were then transferred to 70% glycerol overnight on a shaker at room temperature. The following morning, the co-culture was mounted and imaged using a custom 4-camera spinning disk confocal microscope. The microscope consists of an OpenFrame microscope frame connected to a CrestOptics X-Light V3 spinning disk confocal module that has four Teledyne Photometrics Kinetix cameras mounted to it. It was assembled by Cairn Research UK. All of the organoids were imaged in tiled stacks 800âµm deep using an Olympus Ã10, 0.3âNA air objective with 5âµm zâsteps. The tiles were then stitched using Bigstitcher84 to produce the final image. As the sample holder was transparent on both sides, each organoid was imaged twice, once from each direction.
Image analysis of endothelial cells and SkOs co-cultured with macrophages
Image analysis of endothelial cell culture
To quantify the area covered by endothelial cells in the 2D angiogenesis assay with and without macrophages, phase-contrast images of the wells at 24, 48 and 72âh of culture were analysed using the Fiji distribution of the ImageJ software (v.2.14.0)83. The endothelial area was estimated by measuring the area of the wells covered by all cells (that is, endothelial cells alone or endothelial cells with macrophages). To obtain the endothelial density (in perâcent) the area covered by cells was measured in pixels after segmentation using intensity thresholding and normalized to the total imaged area (constant, 2,115,570 pixels).
Image analysis of SkO culture
To quantify the endothelial area covering organoids with and without macrophages, maximum intensity zâprojections of confocal stacks of CD31+ staining were analysed using the Fiji distribution of the ImageJ software (v.2.14.0)83. The CD31+ endothelial area was measured in µm2 after segmentation using intensity thresholding and normalized to the organoid area (in µm2) to obtain the endothelial density (in perâcent). Each dot represents the endothelial density of one organoid. The analysed stacks contained either 161 or 201 slices, each measuring 1âµm in the zâdimension and up to 1,415âµm by 1,415âµm in the x and y dimensions. Their maximum intensity zâprojections covered a total area ranging from 1.86 to 14.74âmillion of µm2 per organoid.
Whole-mount immunostaining of human prenatal skin sample
For the prenatal tissue specimens, a single PBS rinse was followed by fixation in a freshly prepared 4% PFA solution in 1à PBS at room temperature for half an hour on a shaker. After three PBS washes, specimens were placed in a cold 12.5% SHIELD epoxy solution (LifeCanvas Technologies, SH-Ex) within SHIELD buffer (LifeCanvas Technologies, SH-BS) and gently shaken for 2âdays at 4â°C. Next, specimens were moved to a SHIELD-ON warming solution (LifeCanvas Technologies, SH-ON) for 2âh at 37â°C on a gentle shaker. Following extensive washing in fresh 1à PBS for 8âh (with hourly refreshment) and a 24-h delipidation step at 55â°C in SHIELD Delipidation buffer (LifeCanvas Technologies, DB), specimens were rinsed in room temperature PBST (PBS with 0.1% Triton X-100 and 0.02% sodium azide) for a day. Anti-CD31 (1:100, clone C31.3, Novus Biologicals) and anti-LYVE1 (1:50, polyclonal, Novus Biologicals) primary antibodies were then applied overnight in a 0.1% PBST buffer on a room temperature shaker. Following three 0.1% PBST washes over 3âh, the specimens were incubated for 4âh at room temperature on a shaker with the following secondary antibodies: goat anti-mouse IgG1, Alexa Fluor 488 (ThermoFisher) and goat anti-Rabbit IgG, Alexa Fluor 647 (ThermoFisher). This was then followed by another trio of 0.1% PBST washes. Before imaging, the specimens were conditioned in a 1:1 solution of Easy-Index Matching solution (LifeCanvas Technologies, EI-Z1001) and 1à PBS for 4âh at 37â°C, which was subsequently replaced with a 100% immersion medium for a minimum of 6âh at 37â°C. Imaging was performed using a Nikon A1R HD25 confocal microscope system.
3D rendering
3D volume rendering and segmentation was created using Imarisâ10 software at the Boston Childrenâs Hospital Cellular Imaging Core. For Supplementary Video 1, CD31+ vasculature and LYVE1+ macrophages were processed using the Imaris âSurfacesâ module. Co-localization of the CD31 and LYVE1 channels were processed using the âColocâ feature, generating a separate channel for overlapping signals. The parameters used in the Coloc feature depended on signal overlap and close contact of the CD31 and LYVE1 channels, leading to larger areas labelled as co-localized surfaces than the actual contact points of vessels and macrophages. Classification was based on estimated size and machine learning training. The following build parameters were used for CD31+ endothelial cells: area above 11.0âµm2; for LYVE1+ macrophages: ânumber of voxels Img=1â above 953; for co-localized surfaces: area above 1,004âµm2; filter type âoverlapped volume ratio to surfaces surfaces=LYVE1â threshold=0.00976.
Immunofluorescence of prenatal skin and SkO cryosections
Cryosections (10âµm) were obtained from prenatal skin samples or iPS cell-derived SkOs frozen in OCT (Tissue-Tek OCT). The acquired slides were stored at â80â°C until use. On the day of the experiment, the slides were thawed and dried at room temperature, then fixed for 10âmin in 4% PFA solution in 1à PBS (Alfa Aesar, J61899). Slides were washed with 1à PBS (Gibco, 10010-015) and incubated for 1âh at room temperature with 120âµl per slide of blocking solution (3% goat serum prepared in 1à PBS containing 0.1% Triton X-100 (Millipore, 648466)). A volume of 120âµl per slide of primary antibodies was then applied overnight at 4â°C in the blocking solution (a list of antibodies is supplied in Supplementary Table 38). The following day, the slides were washed 3âtimes with 1à PBS and then incubated for 1â2âh at room temperature with 120âµl per slide of secondary antibodies prepared in blocking solution (Supplementary Table 38). Slides were washed three times with 1à PBS and incubated with 1âµgâmlâ1 of DAPI solution prepared in 1à PBS. Following a final wash with 1à PBS, slides were coverslipped with ProLong Gold Antifade mountant (ThermoFisher, P36930). Slides were dried overnight in the dark at room temperature and imaged using a Leica SP8 Confocal microscope.
Multiplex RNAscope staining and image analysis
Prenatal skin tissue (8, 10 and 15âPCW) was frozen in OCT compound (Tissue-Tek OCT). 4-plex smFISH was performed using a RNAscope Multiplex Fluorescent Detection kit v.2 (ACDBio, 323100) or a RNAscope LS Multiplex Fluorescent Reagent kit v.2 assay and a RNAscope LS 4-Plex Ancillary Kit for LS Multiplex Fluorescent (Advanced Cell Diagnostics (ACD), bio-techne) according to the manufacturerâs instructions. The standard pretreatment for fresh frozen sections of 10â20âμm and permeabilization with Protease IV for 30âmin at room temperature were performed.
Human probes against FOXP3, SHH, SLC26A7, NDP, CDH5, CD68, P2RY12, ACKR3, CXCL12, PDGFD and SERPINB7 transcripts were used (all from ACDBio catalogue probes). Opal dyes (Akoya Biosciences) were used at a dilution of 1:1,000 for the fluorophore step to develop each channel: Opal 520 Reagent Pack (FP1487001KT), Opal 570 Reagent Pack (FP1488001KT) and Opal 650 Reagent Pack (FP1496001KT) and Atto-425. Finally, the slides were counterstained with DAPI and coverslipped for imaging with ProLong Gold Antifade mountant (ThermoFisher, P36930).
4-plex RNAscope slides with FOXP3, SHH, SLC26A7, NDP, CDH5, CD68 and P2RY12 probes were imaged on a Perkin Elmer Opera Phenix Plus High-Content Screening System using a Ã40 (NA 1.1, 0.149âμm per pixel) water-immersion objective with a 2âµm zâstep. The following channels were used: DAPI (excitation (ex.) 375ânm, emission (em.) 435â480ânm); Atto 425 (ex. 425ânm, em. 463â501ânm); Opal 520 (ex. 488ânm, em. 500â550ânm); Opal 570 (ex. 561ânm, em. 570-630ânm); and Opal 650 (ex. 640ânm, em. 650â760ânm). Confocal image stacks were stitched as 2D maximum intensity projections using proprietary Acapella scripts provided by Perkin Elmer and visualized using OMERO Plus (Glencoe Software).
4-plex RNAscope slides with ACKR3, CXCL12, PDGFD and SERPINB7 probes were imaged on the same custom spinning disk confocal microscope used for 3D imaging of the organoids. The objective used was a Ã40 Nikon CFI Plan Apochromat Lambda D (NA 0.95). Imaging was performed with a 1.5âµm zâstep and stitched with the Bigstitcher Fiji plugin to generate a final z-projected image from individual tiles for analysis.
Quantification of FOXP3 coverage was carried out using QuPath image analysis software (v.0.5.1)85. Two-pixel classifiers were trained: one to segment the tissue from the image background and the other to segment out the FOXP3 spots against the background. All of the HF regions were manually segmented out of the whole skin section image. A new segmentation mask was automatically generated from the difference between the whole skin tissue mask and the HF masks. FOXP3 coverage was then calculated separately for the HF regions and the skin tissue by calculating the percentage of the masks that were taken up by segmented FOXP3 spots.
scRNA-seq data analysis
Alignment, quality control, clustering and annotation of prenatal skin dataset
The gene expression data were mapped using CellRanger (v.2.1.1 and v.2.0.2) to an Ensembl 84-based GRCh38 reference (10x Genomicsâdistributed v.1.2.0). The Python package emptydrops (v.0.0.5) was used to detect cells in each sample. Potential doublets were flagged using Scrublet (v.0.2.1)86 as previously described87. Low-quality cells were filtered out first by using a medianâ+â(XâÃâMAD) score (where MAD is the median absolute deviation) of the median score for the mitochondrial UMI fraction (5âÃâMAD), maximum number of UMIs (8âÃâMAD), followed by strict cut-off values (minimum number of genesâ=â200, maximum number of UMIsâ=â50,000, maximum mitochondrial UMI fractionâ=â0.20). Possible maternal contamination (total of 118 cells) was identified using the souporcell pipeline (v.2.4.0)88 as previously described5,58. In brief, samples were pooled on a per-donor basis and processed with souporcell. The common GRCh38 variants file (SNPs with â¥2% frequency from 1k genomes) from souporcell authors was used. The pipeline was run twice, with genotype clusters set to 1 and 2 to obtain models for no maternal contamination and possible maternal contamination. The better model was identified using Bayesian information criterion (BIC), calculated using the formula BICâ=âkn log(m)âââ2l, where k is the number of genotype clusters set for each souporcell run, n denotes the number of loci used for genotype deconvolution, m is the cell count for a given donor, and l is the log likelihood obtained after running the pipeline with each k. The cells with the minor genotype were identified as possible maternal contaminants where identified. Data pre-processing was performed using scanpy (v.1.4.3)89. After pooling data from all samples, genes detected in fewer than three cells were removed, and data were normalized to 1âÃâ104 UMI per cell and log1p transformed.
Highly variable genes were selected on the basis of normalized dispersion (scanpy.pp.highly_variable_genes with flavor = âseuratâ, min_mean = 0.0125, max_mean = 3, min_dispersion = 0.5). Dimensionality reduction was done using principal component analysis and the first 50 principal components were used to compute the nearest-neighbour graph (scanpy.pp.neighbors with n_neighbors = 15). scVI module within scvi-tools (v.0.19.0) was used to correct for donor and 10x kit version batch effects (HVG = 15 000, dropout_rate = 0.2, n_layer = 2)90. Leiden algorithm was used to cluster cells based on the corrected graph with a relatively low resolution (scanpy.tl.leiden with resolution = 0.3) into coarse clusters that were manually annotated into broad lineages using known marker genes.
For each broad lineage, the data were re-processed starting from highly variable gene selection to better reveal finer heterogeneity. At this level, we used Harmony (v.0.0.5)91 and scVI from scvi-tools (v.0.19.0) in parallel for batch correction (again treating each donor as a separate batch) for every broad lineage and observed highly consistent embedding and clustering (data provided on the portal). Leiden clusters at the highest resolution were manually annotated using marker genes identified through the literature search, and their expression of distinctive DEGs specific to each cluster, such as WNT2 expression in WNT2+ fibroblasts. The full list of DEGs for each cluster is provided in Supplementary Table 3. DEGs were calculated using the sctk (Single Cell analysis Tool kit) package (https://github.com/Teichlab/sctk), where filtering is carried out followed by a two-sided Wilcoxon rank-sum test using pass-filter genes only in a one-versus-all fashion. The sctk package also carries out comparisons between the group of interest (one with highest expression) and the next group (second highly expressed), where the maximum proportion of cells expressing the gene in question in the second most highly expressed group was 0.2. For epidermal annotations, we created a combined embedding of prenatal skin and SkO data1, integrated using the Harmony pipeline, as well as integration with adult HF to check annotations, as described below. Harmony-corrected principal components were used to compute the batch-corrected nearest neighbourhood graph, and the Leiden algorithm was used to cluster the integrated data. The sctk package was then used to derive DEGs for each Leiden cluster. Annotation was carried out on the clusters based on marker genes and refined annotations in the SkO data1.
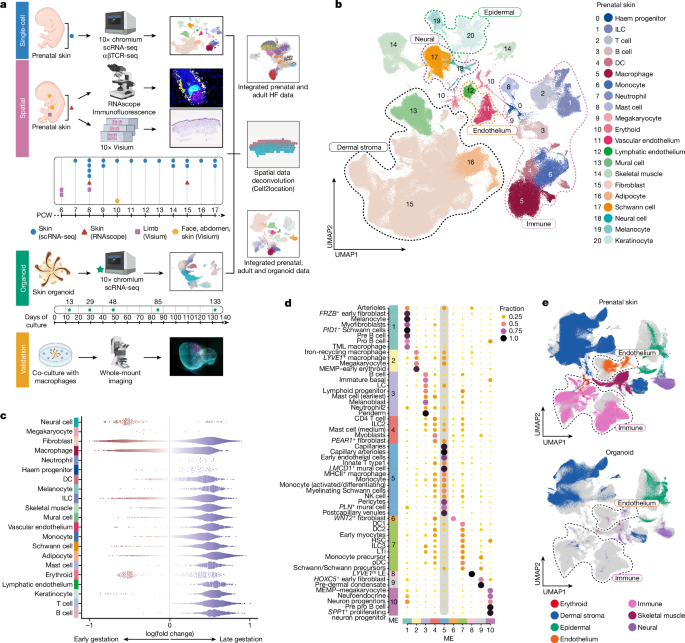
Clusters of doublets were manually flagged and removed, taking into account markers genes and previously calculated scrublet scores. To have a final global visualization of the atlas, a doublet-free UMAP was generated (Fig. 1b).
Processing, clustering and annotation of SkO dataset
Organoid data were pre-processed, filtered, clustered and annotated separately before integration with prenatal skin. In brief, cells filtered by CellRanger (CellRanger 2.1.0 with GRCh38-1.2.0 and CellRanger 3.0.2 with GRCh38-3.0.0) from SkO samples (2 strains, each with 4 time points) were pooled and quality control thresholds for UMI counts, gene counts, percentage of mitochondrial genes and top 50 highly expressed genes were established by fitting Gaussian mixture models to the distribution of each metric respectively. The following thresholds were used: minimum number of genesâ=â450, maximum number of genesâ=â5,731, minimum number of UMIsâ=â1,063, maximum number of UMIsâ=â25,559, maximum mitochondrial UMI fractionâ=â0.133, minimum cumulative percentage of counts for 50 most expressed genes in a cellâ=â23.7%, maximum cumulative percentage of counts for 50 most expressed genes in a cellâ=â56.6%. Highly variable gene selection, dimensionality reduction and KNN graph construction were done using the same method and parameters as prenatal skin. BBKNN (v.1.3.390)92 was used for batch-correction treating combinations of strains and 10x kit versions as batches. Broad lineages were annotated based on known markers. Each broad lineage was then re-processed in the same way as prenatal skin to annotate cell types at higher resolution.
Integration of prenatal skin and SkO datasets
Prenatal skin cells and organoid cells were integrated using Harmony (v.0.0.5)91, treating datasets as batches (prenatal skin or organoid) and within dataset batches as covariates (donor for prenatal skin, strain for SkO, and 10x kit version for both datasets). Leiden clusters were annotated using known markers.
Comparison of prenatal skin, adult skin and SkO datasets: distance-based analysis
Prenatal skin, adult skin and SkO cells were integrated using Harmony (v.0.0.5)91, treating datasets as batches and within-dataset batches as covariates (donor for prenatal and adult skin and strain for organoid, 10x kit version for all datasets). The principal component vectors of the downsampled Harmony-integrated object were then used to transform the gene expression matrix (NumPy (v.1.23.4) function âlinalg.lstsqâ, rcond = âwarnâ) of all cells in the non-downsampled pooled data and project for UMAP visualization (Fig. 1e and Extended Data Fig. 2a). The median transformed gene expression was used to compute the Euclidean distance between prenatal skin, adult skin and SkO for each broad cell cluster, using âspatial.distance_matrixâ function in SciPy (v.1.9.3), which was then plotted as a heatmap (Extended Data Fig. 2c).
Time-encoded cell state predictions: prenatal skin, adult skin and SkO datasets
The median probability of class correspondence between gene expression matrices in single-cell datasets was carried out using a logistic regression (LR) framework as previously described93, based on a similar workflow to CellTypist tool94. Annotated raw scRNA-seq datasets (prenatal skin, adult skin and SkO) were first concatenated, normalized and log-transformed. Linear variational autoencoder (VAE) latent representations were computed using the LDVAE module within scvi-tools (hidden layers = 256, dropout-rate = 0.2, reconstruction-lossâ=ânegative binomial) with dataset and chemistry information taken as technical covariates. ElasticNet LR models were built using the linear_model.LogisticRegression module in the sklearn package (v.0.22). The models were trained on SCVI batch-corrected low-dimensional LDVAE representation of the training data (prenatal and adult skin) using time-encoded labels (age_cell category). Regularization parameters (L1-ratio and alpha) were tuned using the GridSearchCV function in sklearn (v.1.1.3). The test grid was designed with five l1_ratio intervals (0.05, 0.2, 0.4, 0.6 and 0.8), five alpha (inverse of regularization strength) intervals (0.05, 0.2, 0.4, 0.6 and 0.8) at five trainâtest splits and three repeats for cross-validation. The unweighted mean over the weighted mean squared errors (MSEs) of each test fold (the cross-validated MSE) was used to determine the optimal model. The resultant model was used to predict the probability of correspondence between trained time-encoded labels and pre-annotated time_encoded clusters (week of culture_cell category) in the target dataset (SkO). The median probability of training label assignments per predesignated class overall (all cell groups) and for individual broad cell categories were computed (Supplementary Table 5). For visualization, the adult skin dataset was randomly downsampled to 10% (overall or by cell lineage) and resultant LR probabilistic relationship between labels of the training and target datasets were plotted as heatmaps (Extended Data Fig. 2d).
Differential abundance analysis
Differences in cell abundance associated with gestational age were tested using Milo (v.1.0.0)95, correcting for CD45+ and CD45â FACS isolation strategies. We first re-embedded cells into a batch-corrected latent space with a dimension of 30 using scVI model as implemented in scvi-tools considering donor and chemistry as batches. The model was trained using the 15,000 most highly variable genes that were selected based on dispersion (min_meanâ=â0.001, max_meanâ=â10) as previously described58. Where FACS correction was applied, we calculated a FACS isolation correction factor for each sample s sorted with gate i as (fsâ=âlog(piS/Si)) where pi is the true proportion of cells from gate i and S represents the total number of cells from both gates58. A KNN graph of cells was constructed based on distances in the latent space and cells assigned to neighbourhoods using the milopy.core.make_nhoods function (prop = 0.1). The number of cells of each cell type was then counted in each neighbourhood (milopy.core.count_nhoods). Labels were assigned to each neighbourhood based on majority voting (milopy.utils.annotate_nhoods) of cell labels by frequency (>50%). To test for differential abundance across gestational age, prenatal skin data were split into 4 age bins (7â8âPCW, 9â10âPCW, 11â13âPCW and 15â17âPCW), and cell counts were modelled using a negative binomial generalized model, with BenjaminiâHochberg weighted correction as previously described5,58, to test the effects of age (age bins) together with cell sorting correction (milopy.core.DA_nhoods). Significantly differentially abundant neighbourhoods were detected using (SpatialFDRâ<â0.1, log(fold change)â<â0) for early enriched neighbourhoods and (SpatialFDRâ<â0.1, log(fold change)â>â0) for late neighborhoods (Supplementary Table 4).
Cell state predictions: adult HFs, embryonic macrophages, blood vessel organoid, reindeer skin
To compare prenatal skin cells with external datasets (adult HF, embryonic macrophages, blood vessel organoid, reindeer skin)11,57,64,71, the datasets were downsampled to have roughly balanced cell counts per annotated cell type before integration with Harmony (v.0.0.5)91, treating datasets as batches and within dataset batches as covariates (donor for prenatal skin, site for embryonic macrophages, group (cell line: day of culture) for blood vessel organoid, chemistry for reindeer skin).
Comparison of cell type correspondence between datasets and probability prediction was carried out using a LR framework similar to the CellTypist package94. A model was built using the implementation of the linear_model.LogisticRegression module from sklearn package (v.1.1.3) (parameters: penalty: L2, solver: saga, regularization strength Câ=â0.1) and trained on the gene expression matrix of the training dataset using all genes that passed quality control. The resulting model was used to predict the classes in the target dataset. The correspondence between predicted and original classes in the target dataset was computed as the Jaccard index and median probability predictions and visualized as heatmaps. For comparison of the prenatal skin macrophages with embryonic macrophages, the embryonic macrophage dataset was used as training data and prenatal skin macrophages as query; for comparison of the blood vessel organoid with prenatal skin, the prenatal skin dataset (downsampled to maximum of 500 cells per cell type) was used as training data and the blood vessel organoid data as query; for comparison of HF data, merged prenatal and organoid data were used as training data and adult dataset as query; for comparison of the prenatal skin fibroblasts and macrophages with reindeer skin fibroblasts and macrophages, the reindeer skin data subsets were used as training data and prenatal skin data subsets as query.
Cross-species comparison: prenatal skin and mouse skin datasets
The median probability of class correspondence between human and mouse skin single cell datasets was carried out using a LR framework as previously described93, based on a similar workflow to the CellTypist tool94. Annotated raw scRNA-seq datasets (human prenatal skin and mouse embryonic skin41) were first concatenated, normalized, log-transformed and subsetted to retain the top 5,000 highly variable genes (batch_key=dataset) by dispersion. VAE latent representations were computed using scvi-tools (max epochsâ=â400, batch sizeâ=â512) with species, dataset and chemistry information taken as categorical covariates. ElasticNet LR models were built using the linear_model.LogisticRegression module in the sklearn package (v.0.22). The models were trained on SCVI batch-corrected low-dimensional VAE representation of the training data (prenatal skin) for broad cell groupings and refined cell annotations. The resultant models were used to predict the probability of correspondence between trained prenatal skin labels and pre-annotated clusters (broad cell groupings and refined annotations) in the target mouse skin data. The median probability of training label assignments was computed (Supplementary Tables 10 and 11). For visualization, resultant LR probabilistic relationship between labels of the training and target datasets were plotted as heatmaps.
FRZB comparison across developing organs
To compare gene expression of FRZB in fibroblasts across developing organs, a scRNA-seq stromal dataset from multiple developing organs5 (available from the Human Developmental Cell Atlas (https://developmental.cellatlas.io/fetal-immune)) was used, which also includes our prenatal skin scRNA-seq data. The data were normalized to 1âÃâ104 counts per cell (scanpy.pp.normalize_total), log1p transformed (scanpy.pp.log1p) and subsetted to fibroblast cell types only to plot expression of FRZB by organ across gestation time.
Trajectory analysis
The CellRank package96 (v.1.5.2) was used to define cell transition matrices, lineage drivers and rank fate probabilities of terminal state transitions across annotated lineages in a combined embedding of prenatal skin and SkO for keratinocytes and fibroblasts and in the prenatal skin for endothelial cells. Using pp.moments (n_pcs=10, n_neighbours=30) from the scVelo package (v.0.3.0), first order kinetics matrices were imputed. Using the palantir kernel and the velocity kernel in CellRank96, a mixed probability transition matrix was computed with the palantir kernel weighing 70% and the velocity kernel 30%. Schur matrix Eigen decomposition (n_components=25, method=âbrandtsâ) identified macrostates, terminal stages and initial stages. Lineage drivers were then computed for each state using compute_lineage_drivers from CellRank and pseudotime and latent time computed in scVelo (Supplementary Table 7).
In vivoâin vitro trajectory alignment analysis
We used Dynamic Programming-based alignment to evaluate agreement between the single-cell trajectories of prenatal skin and SkO fibroblasts, which describe the in vivo and in vitro differentiation lineages from HOXC5+ early fibroblasts to the Dp. Genes2Genes (G2G)39 is a Bayesian Information-theoretic Dynamic Programming framework that consistently captures matches and mismatches between two trajectories at both the gene level and the cell level. G2G outputs an optimal trajectory alignment that describes a nonlinear mapping of in vivo and in vitro pseudotime points in sequential order. This is based on the cost of matching or mismatching the gene expression distributions of each pair of organoidâreference time points, computed as a statistic of entropy difference between the two hypotheses under the minimum message length97 criterion. This statistic is a Shannon information distance, calculated in the unit of information, nits. Given any gene set, G2G runs Dynamic Programming alignment for each gene, outputting a five-state alignment string over matches (one-to-one/one-to-many/many-to-one) and mismatches (insertions and deletionsâgaps) between the in vivo and in vitro pseudotime points in sequential order, which is analogous to a DNA/protein alignment output. It then computes an alignment similarity measure (that is, the percentage of matches across the alignment string) for each gene (Supplementary Table 9). These alignment strings enabled us to identify different clusters of genes with different alignment patterns. G2G also generates an aggregated cell-level alignment across all gene-level alignments, resulting in an overall alignment similarity measure as well. This aggregated alignment reflects whether the time points are matched or mismatched across genes on average (that is, if there is a higher proportion of gene-level alignments at which a specific time point pair between the two trajectories is matched, then the average alignment includes them as a match).
Using G2G, we examined the in vivo reference versus in vitro query alignment in terms of 1,369 human TFs98. These TFs were taken after filtering zero expressed genes and genes expressed in fewer than ten cells. Given the reference and organoid log1p normalized gene expression matrices of cells and their pseudotime estimates computed using CellRank96, G2G generated fully descriptive TF-level alignments, as well as an aggregated cell-level alignment across those TF-level alignments. Before alignment, the reference and organoid trajectories were discretized over the pseudotime axis in equal length intervals. Note that the number of discrete pseudotime points was determined as 15 based on the optimal binning structures predicted over their pseudotime estimates distributions using the OptBinning99 python package. Also note that at each alignment, these are the discrete time points that are getting matched or mismatched. For each discrete time point of a TF trajectory in a single dataset, G2G estimates its expression distribution as a Gaussian, with a weighted mean and weighted variance computed using all the cells (that is, a Gaussian-kernel-weighted interpolation approach for each cellâs contribution towards this estimation is based on their distance to the particular time point). Next, after interpolating both reference and organoid trajectories using the 15 discrete time points, the Dynamic Programming alignment was run for each TF, and the TF clusters of different alignment patterns (that is, early mismatches, mid mismatches, late mismatches, and complete mismatches) were identified using the G2G function that runs agglomerative hierarchical clustering over the TF-level alignment outputs.
Cellâcell interaction analysis
CellPhoneDB (v.3.0.0) package100 was used to infer cellâcell interactions within the prenatal skin scRNA-seq dataset overall and in early/late gestation and within the SkO scRNA-seq dataset overall. In the overall analysis, we randomly subsampled each cell type into no more than 200 cells 3 separate times, using all cells if a population contained fewer than 200 cells and excluding populations with fewer than 10 cells. The subsampled dataset were analysed separately using the permutation-based method to establish significance (Pâvalue cut-offâ=â0.05). For the analysis by early/late gestation, the prenatal skin scRNA-seq dataset was first split into early (â¤11âPCW) and late (â¥12âPCW) gestation datasets, which were then subsampled (no more than 200 cells per cell type) and individually analysed (Pâvalue cut-offâ=â0.05). A summary output file was created for each analysis run, compiling the interactions for each cell pair (Pâ<â0.05) and adjusting Pâvalues for multiple testing (FDR set at 0.05) (Supplementary Tables 8 and 28). Circos plots (Circlize package (v.0.4.15)101) were used for downstream visualizations of selected significant (adjusted Pâvalueâ<â0.05) interactions between co-locating cell types for interactions found in all subsampled analyses.
To explore inferred interactions between macrophage subsets and endothelial cells (Extended Data Fig. 9a), we aggregated the interactions predicted for each macrophage subset and the different subtypes of endothelial cells (early endothelial cells, arterioles, capillary arterioles, capillaries, postcapillary venules and venules) by averaging the means and using the minimum of the adjusted Pâvalues as previously described5. A curated list of aggregated interactions were plotted for visualization using ggplot2 (v.3.3.6). A similar approach was adopted for assessing interactions between HF dermal and epidermal cells in prenatal skin: for each subset of hair follicle dermal cells, the interactions with early epithelial cells (â¤11âPCW; immature basal) or late epithelial cells (â¥12âPCW; DPYSL2+ basal, POSTN+ basal, placode, matrix, ORS, CL, IRS, cuticle/cortex) were aggregated, and the top 10 interactions per cell pair visualized using a heatmap (Fig. 2g). The same analysis was performed to obtain the top 10 interactions in SkO HFs (Extended Data Fig. 5c), defining early/late to match corresponding cell states as in prenatal skin. The topâ10 interactions identified in prenatal skin HFs were also plotted within the SkO data to highlight similarities and differences between the two (Fig. 2g).
Comparison with adult fibroblasts
Integrated scRNA-seq data from prenatal skin and adult healthy skin (with original annotations)10 was subsetted to the cell group of interest (fibroblasts). DEGs between the adult and prenatal skin fibroblasts were derived using the Wilcoxon rank-sum test implementation in scanpy and adjusted for multiple testing using the BenjaminiâHochberg method (scanpy.tl.rank_genes_groups, method = âwilcoxonâ, corr_method = âbenjamini-hochbergâ). Gene set enrichment analysis was performed using the top 1,000 genes in each group ranked by scores (Supplementary Table 13) and the implementation of the Enrichr workflow102 in the Python package GSEApy (https://gseapy.readthedocs.io/; v.0.10.7) with Gene Ontology Biological Process (2021) as the query database (Supplementary Tables 14, 15). A selected list of genes was plotted to highlight differences between prenatal and adult skin fibroblasts.
Gene set enrichment analysis
To determine the significantly overexpressed genes for gene set enrichment analysis, we first identified the DEGs between cell types for each cell group of interest (myeloid cells) using the Wilcoxon rank-sum test implementation in Scanpy (scanpy.tl.rank_genes_groups, method = âwilcoxonâ). Genes with differential expression log(fold change)â>â1.5 and adjusted Pâvalueâ<â0.01 were considered as significantly overexpressed (Supplementary Table 22). Gene set enrichment analysis was performed using the implementation of the Enrichr workflow102 in the Python package GSEApy (https://gseapy.readthedocs.io/; v.0.10.7) with Gene Ontology Biological Process (2023) and Molecular Signatures Database (MSigDB) Hallmark (2020) as query databases (Supplementary Tables 23â26).
For comparison between early and late cell states, for cell types of interest (WNT2+ fibroblast), we first identified the index cells belonging to early neighbourhoods (SpatialFDRâ<â0.1, log(fold change)â<â0) and late neighbourhoods (SpatialFDRâ<â0.1, log(fold change)â>â0) based on Milo95 differential abundance testing as described above (Supplementary Table 4). DEGs between early and late cell states were computed using the Wilcoxon rank-sum test implementation in scanpy (scanpy.tl.rank_genes_groups, method = âwilcoxonâ). Genes with differential expression log(fold change)â>â1 and adjusted Pâvalueâ<â0.01 were considered as significantly overexpressed (Supplementary Table 16) for gene set enrichment analysis using GSEApy (https://gseapy.readthedocs.io/; v.0.10.7), with Gene Ontology Biological Process (2023) as query the database (Supplementary Tables 17 and 18).
Gene module scoring
Gene module scoring was performed using the sc.tl.score_genes function in scanpy. For angiogenesis gene modules, pre-defined gene sets from the Gene Ontology Biological Process Database (2021) in Enrichr libraries103 were used (downloaded from Enrichr (https://maayanlab.cloud/Enrichr/#libraries)). For endothelial cell modules, gene sets defining tip, stalk, arteriole, venule and lymphatic, capillary (Extended Data Fig. 10g,h) were derived from published literature73,104,105, and for the hypoxia score, a hallmark hypoxia gene list was used. The list of genes for each gene module is provided in Supplementary Tables 27 and 29. The score for each module is the average expression of the gene set provided subtracted with the average expression of a reference set of genes. The reference set comprised 100 genes (ctrl_size=100), which were randomly sampled from all genes in the dataset (default gene_pool) with 25 expression level bins (n_bins=25) used for sampling. For angiogenesis modules, the mean module scores were computed for each cell type of interest (for example, LYVE1+ macrophage) and zâscore normalized for visualization.
Gene regulatory network analysis
The PySCENIC package106 (v.0.11.2) and pipeline were used to identify TFs and their target genes in the combined prenatal skin and SkO scRNA-seq datasets. The ranking database (hg38 refseq-r80 500bp_up_and_100bp_down_tss.mc9nr.feather), motif annotation database (motifs-v9-nr.hgnc-m0.001-o0.0.tbl) were downloaded from the Aertâs laboratory GitHub page. The tool was run 10 times, with a dataset comprising at most 1,000 cells per cell typeâÃâtissue pair (where tissue is prenatal skin or SkO). For each run, an adjacency matrix of TFs and their targets was generated and pruned using the Aertâs group suggested parameters. Only regulons present in at least 6 out of 10 runs were used in the analysis. PySCENIC was used to calculate the regulon specificity score for each cell typeâÃâtissue pair using the aucell function. An average was computed over the multiple runs. These average scores were used to compare regulon activity between prenatal skin and SkO. A gene interaction network was first built by querying the STRING database with GATA2 target genes, then pruned to only keep genes reported as associated with GATA2. The list was further truncated to 12 genes, by keeping genes that met the following criteria: (1) TFs in the five most active regulons detected in fetal skin; and/or (2) organoid capillary arterioles; and/or (3) associated with pseudotime (that is, in trajectories); and/or (4) VEGF receptors; and/or (5) in the selected gene ontology terms chosen for their role in angiogenesis, extracellular matrix organization, or cell migration, communication, proliferation, or death (GO:0045765, GO:0001568, GO:0030334, GO:0010646, GO:0001936, GO:0045446, GO:0002040, GO:0030155, GO:0010941 and GO:0030198).
Comparison of pro-angiogenic and anti-angiogenic genes between prenatal skin and SkO
The prenatal skin and SkO datasets were integrated using Harmony (v.0.0.5)91 as described above. Differential expression analysis was performed between prenatal skin and SkO cells (all cell types) using the standard scanpy workflow (scanpy.tl.rank_genes_groups, method = âwilcoxonâ). Identified DEGs were filtered to only retain those coding for secreted proteins107 (Supplementary Table 39). Gene set enrichment analysis was performed on downregulated and upregulated genes separately, using the implementation of the Enrichr workflow102 in the Python package GSEApy (https://gseapy.readthedocs.io/) with Gene Ontology Biological Process (2021) as the query database. Significant gene ontology terms (adjusted Pâvalueâ<â0.05) (Supplementary Tables 33 and 34) were filtered based on their relevance to vasculature. Only DEGs involved in pathways thereby selected were chosen and their role in prenatal skin angiogenesis checked in the literature.
NicheNet analysis
We used NicheNet74 (v.1.1.1) to infer ligandâtarget gene links by combining scRNA-seq data (prenatal skin and SkO) of interacting cells (sender and receiver cells) with existing knowledge on signalling and gene regulatory networks. An open-source R implementation including integrated data sources used in the analysis are available at GitHub (https://github.com/saeyslab/nichenetr). NicheNetâs ligandâactivity analysis first assesses and ranks ligands in the sender cell type (macrophage subsets), which best predict observed changes in expression of target genes of interest in the receiver cell types (endothelial cells) compared with background genes. Potential ligands were defined as all ligands in the NicheNet model that were expressed in at least 10% of cells in each macrophage (sender) cluster and had at least one specific receptor expressed in at least 10% of endothelial (receiver) cells. Target genes of interest were identified as DEGs between conditions (prenatal skin versus SkO) in receiver cells using FindMarkers function in NicheNet (adjusted Pâvalueââ¤â0.05 and average log2(fold change)â>â0.25, expressed in at least 10% of endothelial cells). Background genes were all genes in the NicheNet model that were expressed in at least 10% of receiver cells.
Ligands were prioritized based on ligand activity scores, calculated as the Pearson correlation coefficient between a ligandâs target predictions and the observed target gene expression (Supplementary Table 35). The topâ20 ligands were used to predict active target genes (top 200 overall) and construct the active ligandâtarget links (Supplementary Table 36). Receptors of the top-ranked ligands were identified from the NicheNet model, which filters for only bona fide ligandâreceptor interactions documented in the literature and publicly available databases (Supplementary Table 37). To infer signalling paths, we defined our ligand (VEGFA, in red) and target genes (GATA2, in blue) of interest. NicheNet identifies which TFs best regulate the target genes and are most closely downstream of the ligand based on weights of the edges in its integrated ligand signalling and gene regulatory networks. The shortest paths between these TFs and the defined ligand are selected and genes along these paths are considered as relevant signalling mediators (in grey). Signalling mediators are prioritized by cross-checking the interactions between the defined ligand, target genes and identified TFs and signalling mediators across the different integrated data sources in NicheNet.
Spatial data analysis
Spatial transcriptomics data were mapped using Space Ranger (v.2.0.1) using GRCh38-1.2.0 reference. In parallel, we manually selected skin-overlapping spots in embryonic limb data12, comprising samples from the following ages: 6âPCW (nâ=â2, replicateâ=â2 each) and 8âPCW (nâ=â1, replicateâ=â3). To map cell types identified by scRNA-seq in the profiled spatial transcriptomics slides, we used the Cell2location (v.0.1) method15. First, we trained a negative binomial regression model to estimate reference transcriptomic profiles for all the cell types profiled with scRNA-seq in the organ (nâ=â15 samples). We excluded very lowly expressed genes using the filtering strategy recommended by the authors of Cell2location (cell_count_cutoff=5, cell_percentage_cutoff2=0.03, nonz_mean_cutoff=1.12). Cell types for which fewer than 20 cells were identified in samples â¤10âPCW were excluded from the reference. Individual 10x samples were considered as a batch, donor and chemistry type information was included as categorical covariate. Training was performed for 250 epochs and reached convergence according to manual inspection. Next, we estimated the abundance of cell types in the spatial transcriptomics slides using reference transcriptomic profiles of different cell types. All slides were analysed jointly. The following Cell2location hyperparameters were used: (1) expected cell abundance (N_cells_per_location)â=â30; (2) regularization strength of detection efficiency effect (detection_alpha)â=â20. The training was stopped after 50,000 iterations. All other parameters were used at default settings. Cell2location estimates the posterior distribution of cell abundance of every cell type in every spot. Posterior distribution was summarized as 5% quantile, representing the value of cell abundance that the model has high confidence in, and therefore incorporating the uncertainty in the estimate into values reported in the paper and used for downstream co-location analysis.
To identify microenvironments of co-locating cell types, we used NMF. We first normalized the matrix of estimated cell type abundances by dividing it by per-spot total abundances. Resulting matrix Xn of dimensions nâÃâc, where n is the total number of spots in the Visium slides and c is the number of cell types in the reference was decomposed as Xnâ=âWZ, where W is a nâÃâd matrix of latent factor values for each spot and Z is a dâÃâc matrix representing the fraction of abundance of each cell type attributed to each latent factor. Here latent factors correspond to tissue microenvironments defined by a set of co-localized cell types. We use the NMF package for R108, setting the number of factors dâ=â10 and using the default algorithm109. NMF coefficients were normalized by a per-factor maximum. We ran NMF 100 times and constructed the coincidence matrix. Then we selected the best run based on the lower mean silhouette score calculated on the coincidence matrix. If more than one run had the minimal mean silhouette, we selected one with smaller deviance (as reported by NMF function).
For cell type abundance correlation analysis, we used a per-spot normalized Xn matrix. Pearson correlation coefficient was calculated for each pair of cell types (all possible pairs computed) and each sample. For visualization of correlation analysis, selected cell pairs were plotted, guided by NMF results and which cell groups or categories formed microenvironments. For example, macrophages formed microenvironments with endothelial cells (ME1 and ME5), with neural cells (ME1 and ME5) and fibroblasts (ME1, ME4 and ME5) in Fig. 1d (Supplementary Table 40).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.