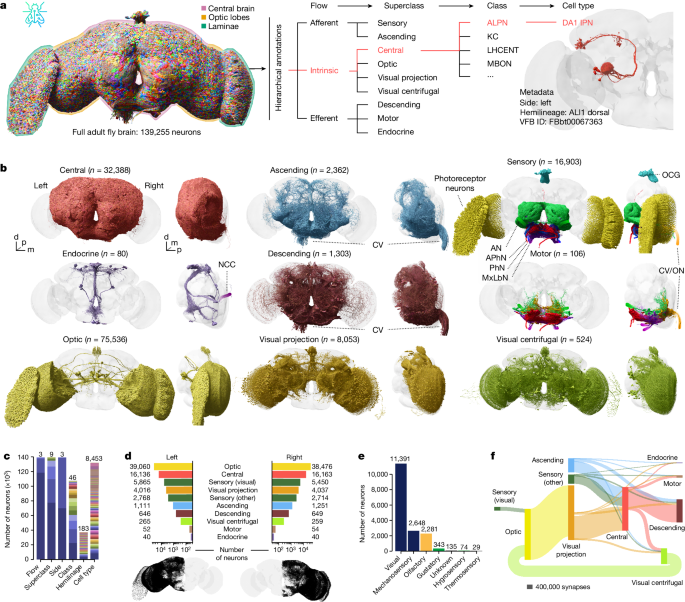
Annotations
Base annotations
At the time of writing, the general FlyWire annotation system operates in a read-only mode in which users can add additional annotations for a neuron but cannot edit or delete existing annotations. Furthermore, the annotations consist of a single free-form text field bound to a spatial location. This enabled many FlyWire users (including our own group) to contribute a wide range of community annotations, which are reported in our companion paper1 but are not considered in this study. As it became apparent that a complete connectome could be obtained, we found that this approach was not a good fit for our goal of obtaining a structured, systematic and canonical set of annotations for each neuron with extensive manual curation. We therefore set up a web database (seatable; https://seatable.io/) that allowed records for each neuron to be edited and corrected over time; columns with specific acceptable values were added as necessary.
Each neuron was defined by a single point location (also known as a root point) and its associated PyChunkedGraph supervoxel. Root IDs were updated every 30âmin by a Python script based on the fafbseg package (Table 1) to account for any edits. The canonical point for the neuron was either a location on a large-calibre neurite within the main arbour of the neuron, a location on the cell body fibre close to where it entered the neuropil or a position within the nucleus as defined by the nucleus segmentation table80. The former was preferred as segmentation errors in the cell body fibre tracts regularly resulted in the wrong soma being attached to a given neuronal arbour. These soma swap errors persisted late into proofreading and, when fixed, resulted in annotation information being attached to the wrong neuron until this in turn was fixed.
We also note that our annotations include a number of non-neuronal cells/objects such as glia cells, trachea and extracellular matrix that others might find useful (superclass not_a_neuron; listed in Supplementary Data 2).
Soma position and side
Besides the canonical root point, the soma position was recorded for all neurons with a cell body. This was either based on curating entries in the nucleus segmentation table (removing duplicates or positions outside the nucleus) or on selecting a location, especially when the cell body fibre was truncated and no soma could be identified in the dataset. These soma locations were critical for a number of analyses and also allowed a consistent side to be defined for each neuron. This was initialized by mapping all soma positions to the symmetric JRC2018F template and then using a cutting plane at the midline perpendicular to the mediolateral (x) axis to define left and right. However, all soma positions within 20âµm of the midline plane were then manually reviewed. The goal was to define a consistent logical soma side based on examination of the cell body fibre tracts entering the brain; this ultimately ensured that cell types present, for example, in one copy per brain hemisphere, were always annotated so that one neuron was identified as the left and the other the right. In a small number of cases, for example, for the bilaterally symmetric octopaminergic ventral unpaired medial neurons, we assigned side as âcentralâ.
For sensory neurons, side refers to whether they enter the brain through the left or the right nerve. In a small number of cases we could not unambiguously identify the nerve entry side and assigned side as ânaâ.
Biological outliers and sample artefacts
Throughout our proofreading, matching and cell typing efforts, we recorded cases of neurons that we considered to be biological outliers or showed signs of sample preparation and/or imaging artefacts.
Biological outliers range from small additional/missing branches to entire misguided neurite tracks, and were typically assessed within the context of a given cell type and best possible contralateral matches within FlyWire and/or the hemibrain. When biological outliers were suspected, careful proofreading was undertaken to avoid erroneous merges or splits of neuron segmentation.
Sample artefacts come in two flavours:
(1) A small number of neurons exhibit a dark, almost black cytosol, which caused issues in the segmentation as well as synapse detection. This effect is often restricted to the neuronsâ axons. We consider these sample artefacts because it is not always consistent within cell types. For example, the cytosol in the axons of DM3 adPN is dark on the left and normal light on the right. Because the dark cytosol leads to worse synapse detection, probably due to lower contrast between the cytosol and synaptic densities, we typically excluded neurons (or neuron types) with sample artefacts from connectivity analyses. Anecdotally, this appears to happen at a much higher frequency in sensory neurons compared with in brain-intrinsic neurons.
(2) Some neurons are missing large arbours (for example, a whole axon or dendrite) because a main neurite suddenly ends and cannot be traced any further. This typically happens in commissures where many neurites co-fasculate to cross the brainâs midline. In some but not all cases, we were able to bridge those gaps and find the missing branch through leftâright matching. Where neurons remained incomplete, we marked them as outliers.
Whether a neuron represents a biological outlier or exhibits sample preparation/segmentation artefacts is recorded in the status column of our annotations as âoutlier_bioâ and âoutlier_segâ, respectively. Note that these annotations are probably less comprehensive for the optic lobes than for the central brain. Examples plus quantification are presented in Extended Data Fig. 5.
Hierarchical annotations
Hierarchical annotations include flow, superclass, class (plus a subclass field in certain cases) and cell type. The flow and superclass were generally assigned based on an initial semi-automated approach followed by extensive and iterative manual curation. See Supplementary Table 3 for definitions and the sections below for details on certain superclasses.
Based on the superclasses we define two useful groupings which are used throughout the main text:
Central brain neurons consist of all neurons with their somata in the central brain defined by the five superclasses: central, descending, visual centrifugal, motor and endocrine.
Central brain associated neurons further include superclasses: visual projection neurons (VPNs), ascending neurons and sensory neurons (but omit sensory neurons with cell class: visual).
Cell classes in the central brain represent salient groupings/terms that have been previously used in the literature (examples are provided in Supplementary Table 3). For sensory neurons, the class indicates their modality (where known). For optic-lobe-intrinsic neurons cell class indicates their neuropil innervation: for example, cell class âMEâ are medulla local neurons, âLA>MEâ are neurons projecting from the lamina to the medulla and âME>LO.LOPâ are neurons projecting from the medulla to both lobula and lobula plate.
Hemilineage annotations
Central nervous system lineages were initially mapped for the third instar larval brain, where, for each lineage, the neuroblast of origin and its progeny are directly visible81,82,83,84. Genetic tools that allow stochastic clonal analysis85 have enabled researchers to visualize individual lineages as GFP-marked âclonesâ. Clones reveal the stereotyped morphological footprint of a lineage, its overall âprojection envelopeâ32, as well as the cohesive fibre bundlesâhemilineage-associated tracts (HATs)âformed by neurons belonging to it. Using these characteristics, lineages could be also identified in the embryo and early larva86,87, as well as in pupae and adults31,32,33,34,37,88. HATs can be readily identified in the EM image data, and we used them, in conjunction with clonal projection envelopes, to identify hemilineages in the EM dataset through a combination of the following methods:
(1) Visual comparison of HATs formed by reconstructed neurons in the EM, and the light microscopy map reconstructed from anti-Neuroglian-labelled brains31,33,34. In cross-section, tracts typically appear as clusters of 50â100 tightly packed, rounded contours of uniform diameter (~200ânm), surrounded by neuronal cell bodies (when sectioned in the cortex) or irregularly shaped terminal neurite branches and synapses (when sectioned in the neuropil area; Fig. 2c). The point of entry and trajectory of a HAT in the neuropil is characteristic for a hemilineage.
(2) Matching branching pattern of reconstructed neurons with the projection envelope of clones: as expected from the light microscopy map based on anti-Neuroglian-labelled brains31, the majority of hemilineage tracts visible in the EM dataset occur in pairs or small groups (3â5). Within these groups, individual tracts are often lined by fibres of larger (and more variable) diameter, as shown in Fig. 2c. However, the boundary between closely adjacent hemilineage tracts is often difficult to draw based on the EM image alone. In these cases, visual inspection and quantitative comparison of the reconstructed neurons belonging to a hemilineage tract with the projection envelope of the corresponding clone, which can be projected into the EM dataset through Pyroglancer (Table 1), assists in properly assigning neurons to their hemilineages.
(3) Identifying homologous HATs across three different hemispheres (left and right of FlyWire, hemibrain): by comparison of morphology (NBLAST38), as well as connectivity (assuming that homologous neurons share synaptic partners), we were able to assign the large majority of neurons to specific HATs that matched in all three hemispheres.
In the existing literature, two systems for hemilineage nomenclature are used: Ito/Lee33,34 and Hartenstein31,32. Although these systems overlap in large parts, some lineages have been described in only one but not the other nomenclature. In the main text, we provide (hemi)lineages according to the ItoLee nomenclature for simplicity. Below and in the Supplementary Information, we also provide both names as ItoLee/Hartenstein, and the mapping between the two nomenclatures is provided in Supplementary Data 3. From previous literature, we expected a total of around 119 lineages in the central brain, including the gnathal ganglia (GNG)31,32,33,34,84. Indeed, we were able to identify all 119 lineages based on light-level clones and tracts, as well as the HATs in FlyWire. Moreover, we found one lineage, LHp3/CP5, which could not be matched to any clone. Thus, together, we have identified 120 lineages.
By comprehensively inspecting the hemilineage tracts originally in CATMAID and then in FlyWire, we can now reconcile previous reports. Specifically, new to refs. 33,34 (ItoLee nomenclature) are: CREl1/DALv3, LHp3/CP5, DILP/DILP, LALa1/BAlp2, SMPpm1/DPMm2 and VLPl5/BLVa3_or_4âwe gave these neurons lineage names according to the naming scheme in refs. 33,34. New to ref. 31 (Hartenstein nomenclature) are: SLPal5/BLAd5, SLPav3/BLVa2a, LHl3/BLVa2b, SLPpl3/BLVa2c, PBp1/CM6, SLPpl2/CP6, SMPpd2/DPLc6, PSp1/DPMl2 and LHp3/CP5âwe named these units according to the Hartenstein nomenclature naming scheme. We did not take the following clones from ref. 33 into account for the total count of lineages/hemilineages, because they originate in the optic lobe and their neuroblast of origin has not been clearly demonstrated in the larva: VPNd2, VPNd3, VPNd4, VPNp2, VPNp3, VPNp4, VPNv1, VPNv2 and VPNv3.
Notably, although light-level clones from refs. 33,34 match very well the great majority of the time, sometimes clones with the same name only match partially. For example, the AOTUv1_ventral/DALcm2_ventral hemilineage seems to be missing in the AOTUv1/DALcm2 clone in the Ito collection33. There appears to be a similar situation for the DM4/CM4, EBa1/DALv2 and LHl3/BLVa2b lineages. When there is a conflict, we have preferred clones as described in ref. 34.
For calculating the total number of hemilineages, to keep the inclusion criteria consistent with the lineages, we included the type II lineages (DL1-2/CP2-3, DM1-6/DPMm1, DPMpm1, DPMpm2, CM4, CM1, CM3) by counting the number of cell body fibre tracts, acknowledging that they may or may not be hemilineages. Neuroblasts of type II lineages, instead of generating ganglion mother cells that each divide once, amplify their number, generating multiple intermediate progenitors that in turn continue dividing like neuroblasts28,89,90. It has not been established how the tracts visible in type II clones (and included in Extended Data Fig. 3 and Supplementary Data 3 and 4) relate to the (large number of) type II hemilineages.
There are also 3 type I lineages (VPNl&d1/BLAl2, VLPl2/BLAv2 and VLPp&l1/DPLpv) with more than two tracts in the clone; we included these additional tracts in the hemilineages provided in the text. Without taking these type I and type II tracts into account, we identified 141 hemilineages.
A minority of neurons in the central brain could not reliably be assigned to a lineage. These mainly include the (putative) primary neurons (3,780). Primary neurons, born in the embryo and already differentiated in the larva, form small tracts with which the secondary neurons become closely associated91. In the adult brain, morphological criteria that unambiguously differentiate between primary and secondary neurons have not yet been established. In cases in which experimental evidence exists27, primary neurons have significantly larger cell bodies and cell body fibres. Loosely taking these criteria into account we surmise that a fraction of primary neurons forms part of the HATs defined as described above. However, aside from the HATs, we see multiple small bundles, typically close to but not contiguous with the HATs, which we assume to consist of primary neurons. Overall, these small bundles contained 3,780 neurons, designated as primary or putative primary neurons.
Hemilineage annotations in hemibrain
Hemilineage annotations in hemibrain were generated using the hemilineage annotations in FlyWire as the ground truth. For each hemilineage, we first obtained potential hemibrain matches to FlyWire neurons using a combination of NBLAST38 scores and cell body fibre/cell type annotations. We then clustered neurons in all three hemispheres (FlyWire left, FlyWire right, hemibrain potential candidates) by morphology, and went through the clusters, to make sure that the hemilineage annotations correspond across brains at the finest level possible. To ensure that no neurons within a hemilineage were missed, we examined the cell body fibre bundles of each hemilineage in the hemibrain at the EM level. To further guarantee the completeness of hemilineage annotations, we inventoried all right hemisphere neurons in hemibrain with a cell type annotation, to ensure all neurons with a type annotation were assigned a hemilineage annotation where possible.
Morphological groups
Within a hemilineage, subgroups of neurons often share distinctive morphological characteristics. These morphological groups were identified for all hemilineages as follows. Neurons from FlyWire and hemibrain were transformed into the same hemisphere and pairwise NBLAST scores were generated for all neurons within a hemilineage. Intrahemilineage NBLAST scores were then clustered using HDBSCAN92, an adaptive algorithm that does not require a uniform threshold across all clusters, and that does not assume spherical distribution of data points in a cluster, compared to other clustering algorithms such as k-means clustering.
To test the robustness of the morphological groups, we reran the above analysis across one, two or three hemispheres. This treatment sometimes gave slightly different results. However, some groups of neurons consistently co-clustered across the different hemispheres; we termed these âpersistent clustersâ. Early-born neurons, which are often morphologically unique, frequently failed to participate in persistent clusters, and were omitted from further analysis. We linked these persistent clusters across hemispheres using two- and three-hemisphere clustering: for example, when clustering FlyWire left and FlyWire right together for hemilineage AOTUv3_dorsal, the TuBu neurons from both the left and right hemispheres would fall into one cluster, which we termed a morphological group. Morphological groups are therefore defined by consistent across-hemisphere clustering. When neurons of a given hemilineage were sufficiently contained by the hemibrain volume, all three hemispheres (two from FlyWire and one from hemibrain) were used; otherwise, the two hemispheres from FlyWire were used. As we prioritized consistency across 1, 2 and 3 hemisphere clustering, a minority of neurons with a hemilineage annotation do not have a morphological group. For example, if neuron type A clusters with type B in one-hemisphere clustering, but clusters with type C (and not B) in two-hemisphere clustering, then type A will not have a morphological group annotation.
After generating the morphological groups, we cross-checked these annotations against existing cross-identified hemibrain types and (FlyWire only) cell types. In a minority of cases, neurons of one hemibrain/cell type were annotated with multiple morphological groups. This occasionally reflected errors in assigning types, which were corrected; and others where individual neurons from a type were singled out due to additional branches/reconstruction issues. We therefore manually corrected some morphological group annotations to make them correspond maximally with the hemibrain/cell type annotations.
Overall, we divide hemilineages in each hemisphere into 528 morphological groups, with hemilineages typically having 1â6 morphological groups (10/90 quantile) and with each morphological group containing 2â52 neurons in each hemisphere (10/90 quantile).
Cell typing
Using methods described in detail in the sections below, we defined cell types for 96.4% of all neurons in the brainâ98% and 92% for the central brain and optic lobes, respectively. The remaining 3.6% of neurons were largely (1) optic lobe local neurons for which we could not find a prior in existing literature or (2) neurons without clear contralateral pairings, including a number of neurons on the midline.
About 21% of our cell type annotations are principally derived from the hemibrain cell type matching effort (see the section below). The remainder was generated either by comparing to existing literature (for example, in case of optic lobe cell types or sensory neurons) and/or by finding left/right balanced clusters through a combination of NBLAST and connectivity clustering (Fig. 6 and Extended Data Figs. 8 and 9). New types were given a simple numerical cross-brain identifier (for example, CB0001) or, in the case of ascending neurons (ANs)/descending neurons(DNs), a more descriptive identifier (see the section below) as a provisional cell type label. A flow chart summary is provided in Extended Data Fig. 12.
For provenance, we provide two columns of cell types in our Supplementary Data:
hemibrain_type always refers to one or more hemibrain cell types; in rare occasions where a matched hemibrain neuron did not have a type, we recorded body IDs instead.
cell_type contains types that are either not derived from the hemibrain or that represent refinements (for example, a split or retyping) of hemibrain types.
Neurons can have both a cell_type and a hemibrain_type entry, in which case, the cell_type represents a refinement or correction and should take precedence. This generates the reported total count of 8,453 terminal cell types and includes 3,643 hemibrain-derived cell types (Fig. 3h (right side of the flow chart)) and 4,581 proposals for new types. New types consist of 3,504 CBXXXX types, 65 new visual centrifugal neuron types (âcâ prefix, for example, cL08), 173 new VPN types (âeâ suffix, for example, LTe07), 602 new AN types (âAN_â or âSA_â prefix, for example, AN_SMP_1) and 237 new DN types (âeâ suffix, for example, DNge094). The remaining 229 types are cell types known from other literature, for example, columnar cell types of the optic lobes.
Hemibrain cell type matching
We first used NBLAST38 to match FlyWire neurons to hemibrain cell types (see âMorphological comparisonsâ section). From the NBLAST scores, we extracted, for each FlyWire neuron, a list of potential cell type hits using all hits in the 90th percentile. Individual FlyWire neurons were co-visualized with their potential hits in neuroglancer (see the âData availabilityâ and âCode availabilityâ sections) and the correct hit (if found) was recorded. In difficult cases, we would also inspect the subtree of the NBLAST dendrograms containing the neurons in questions to include local cluster structure in the decision making (Extended Data Fig. 4e). In cases in which two or more hemibrain cell types could not be cleanly delineated in FlyWire (that is, there were no corresponding separable clusters) we recorded composite (many:1) type matches (Fig. 3i and Extended Data Figs. 4g and 12).
When a matched type was either missing large parts of its arbours due to truncation in the hemibrain or the comparison with the FlyWire matches suggested closer inspection was required, we used cross-brain connectivity comparisons (see the section below) to decide whether to adjust (split or merge) the type. A merge of two or more hemibrain types was recorded as, for example, SIP078,SIP080, while a split would be recorded as PS090a and PS090b (that is, with a lower-case letter as a suffix). In rare cases in which we were able to find a match for an untyped hemibrain neuron, we would record the hemibrain body ID as hemibrain type and assign a CBXXXX identifier as cell type.
Finally, the hemibrain introduced the concept of morphology types and âconnectivity typesâ2. The latter represent refinements of the former and differ only in their connectivity. For example, morphology type SAD051 splits into two connectivity types: SAD051_a and SAD051_b, for which the _{letter} indicates that these are connectivity types. Throughout our FlyWireâhemibrain matching efforts we found connectivity types hard to reproduce and our default approach was to match only up to the morphology type. In some cases, for example, antennal lobe local neuron types like lLN2P_a and lLN2P_b, we were able to find the corresponding neurons in FlyWire.
Note that, in numerous cases that we reviewed but remain unmatched, we encountered what we call ambiguous âdaisy-chainsâ: imagine four fairly similar cell types, A, B, C and D. Often these adjacent cell types represent a spectrum of morphologies where A is similar to B, B is similar to C and C is similar to D. The problem now is in unambiguously telling A from B, B from C and C from D. But, at the same time, A and D (on the opposite ends of the spectrum) are so dissimilar that we would not expect to assign them the same cell type (Fig. 3k and Extended Data Fig. 4h). These kinds of graded or continuous variation have been observed in a number of locations in the mammalian nervous system and represent one of the classic complications of cell typing18. Absent other compelling information that can clearly separate these groups, the only reasonable option would seem to be to lump them together. As this would erase numerous proposed hemibrain cell types, the de facto standard for the fly brain, we have been conservative about making these changes pending analysis of additional connectome data2.
Hemibrain cell type matching with connectivity
In our hemibrain type matching efforts, about 12% of cell types could not be matched 1:1. In these cases, we used across-dataset connectivity clustering (for example, to confirm the split of a hemibrain type or a merger of multiple cell types). To generate distances, we first produced separate adjacency matrices for each of the three hemispheres (FlyWire left, right and hemibrain). In these matrices, each row is a query neuron and each column is an up- or downstream cell type; the values are the connection weights (that is, number of synapses). We then combine the three matrices along the first axis (rows) and retain only the cell types (columns) that have been cross-identified in all hemispheres. From the resulting observation vector, we calculate a pairwise cosine distance. It is important to note that this connectivity clustering depends absolutely on the existence of a corpus of shared labels between the two datasetsâwithout such shared labels, which were initially defined by morphological matching as described above, connectivity matching cannot function.
This pipeline is implemented in the coconatfly package (Table 1), which provides a streamlined interface to carry out such clustering. For example the following command can be used to see if the types given to a selection of neurons in the Lateral Accessory Lobe (LAL) are robust:
cf_cosine_plot(cf_ids(â/type:LAL0(08|09|10|42)â, datasets=c(âflywireâ,ââhemibrainâ)))
.
An optional interactive mode allows for efficient exploration within a web browser. For further details and examples, see https://natverse.org/coconatfly/.
Defining robust cross-brain cell types
In Fig. 6, we used two kinds of distance metricsâone calculated from connectivity alone (used for FC1â3; Fig. 6eâg) and a second combining morphology + connectivity (used for FB1â9; Fig. 6h and Extended Data Fig. 8bâf) to help define robust cross-brain cell types. The connectivity distance is as described in the âHemibrain cell type matching with connectivityâ section above). We note that the central complex retyping used FlyWire connectivity from the 630 release. The combined morphology + connectivity distances were generated by taking the sum of the connectivity and NBLAST distances. Connectivity-only works well in the case of cell types that do not overlap in space but instead tile a neuropil. For cell types that are expected to overlap in space, we find that adding NBLAST distances is a useful constraint to avoid mixing of otherwise clearly different types. From the distances, we generated a dendrogram representation using the Ward algorithm and then extracted the smallest possible clusters that satisfy two criteria: (1) each cluster must contain neurons from all three hemispheres (hemibrain, FlyWire right and FlyWire left); (2) within each cluster, the number of neurons from each hemisphere must be approximately equal.
We call such clusters âbalancedâ. The resulting groups were then manually reviewed.
Defining new provisional cell types
After the hemibrain type matching effort, around 40% of central brain neurons remained untyped. This included both neurons mostly or entirely outside the hemibrain volume (for example, from the GNG) but also neurons for which the potential hemibrain type matches were too ambiguous. To provide provisional cell types for these neurons, we ran the same cell typing pipeline described in the âDefining robust cross-brain cell typesâ section above on the two hemispheres of FlyWire alone. In brief, we produced a morphology + connectivity co-clustering for each individual hemilineage (neurons without a hemilineage such as putative primary neurons were clustered separately) and extracted âbalancedâ clusters, which were manually reviewed (Fig. 6i,j and Extended Data Fig. 9). Reviewed clusters were then used to add new or refine existing cell and hemibrain types:
-
Clusters consisting entirely of previously untyped neurons were given a provisional CBXXXX cell type.
-
Clusters containing a mix of hemibrain-typed and untyped neurons typically meant that, after further investigation, the untyped neurons were given the same hemibrain type.
-
Hemibrain types split across multiple clusters were double checked (for example, by running a triple-hemisphere connectivity clustering), which often led to a split of the hemibrain type; for example, SMP408 was split into SMP408aâd.
-
In rare cases, clusters contained a mix of two or more hemibrain types; these were double checked and the hemibrain types corrected (for example, by merging two or more hemibrain types, or by removing hemibrain type labels).
To validate a subset of the new, provisional cell types, we re-ran the clustering using three hemispheres (FlyWire + hemibrain) on 25 cross-identified hemilineages that are not truncated in the hemibrain (Extended Data Fig. 9). The procedure was otherwise the same as for the double-clustering.
Optic lobe cell typing
We provide cell type annotations for >92% of neurons in both optic lobes. The vast majority of these types are based on previous literature42,93,94,95,96,97,98,99. We started the typing effort by annotating well-known large tangential cells (for example, Am1 or LPi12), VPNs (for example, LT1s) as well as photoreceptor neurons. From there, we followed two general strategies, sometimes in combination: (1) for neurons with known connectivity fingerprints, we specifically hunted upstream or downstream of neurons of interest (for example, looking for T4a neurons upstream of LPi12). (2) We ran connectivity clustering as described above on both optic lobes combined. Clusters were manually reviewed and matched against literature. This was done iteratively; with each round adding new or refining existing cell types to inform the next round of clustering. Clusters that we could not confidently match against a previously described cell type were assigned a provisional (CBXXXX) type.
This effort was carried out independently of other FlyWire optic lobe intrinsic neuron typing, including ref. 23; the sole exception was the Mi1 cell type, which was initially based on annotations reported previously100 and then reviewed. For this reason ref. 100 should be cited for the Mi1 annotations. Note that our typing focuses on previously reported cell types rather than defining new ones, but covers both optic lobes to enable accurate typing of visual project neurons (by defining their key inputs). For the 38,461 neurons of the right optic lobe (for which a comparison is possible), we report 156 cell types for 35,567 neurons compared with 229 cell types for 37,345 neurons in ref. 23.
VPNs and VCNs
Similar to cell typing in the central brain, a significant proportion of VPN (61%) and visual centrifugal neuron (VCN) (60%) types are derived from the hemibrain (see the âHemibrain cell type matchingâ section). These annotations are listed in the hemibrain_type column in the Supplementary Data.
To assign cell types to the remaining neurons and in some cases also to refine existing hemibrain types, we ran a double-hemisphere (FlyWire leftâright) co-clustering. For VCNs, this was done as part of the per-hemilineage morphology-connectivity clustering described in the âDefining new provisional cell typesâ section above. For VPNs of which the dendrites typically tile the optic neuropils, we generated and reviewed a separate connectivity-only clustering on all VPNs together. Groups extracted from this clustering were also cross-referenced with new literature from parallel typing efforts100,101 and those new cell type names were preferred for the convenience of the research community. In cases in which literature references could not be found, systematic names were generated de novo using the schemata below.
For VPNs the nomenclature follows the format [neuropil][C/T][e][XX], where neuropil refers to regions innervated by VPN dendrites; C/T denotes columnar versus tangential organization; e indicates identification through EM; and XX represents a zero padded two digit number.
For example: âMTe47â for âmedulla-tangential 47â.
For VCNs, the nomenclature follows the format [c][neuropil][XX], where c denotes centrifugal; neuropil refers to regions innervated by VCN axons; and XX represents a zero padded two digit number.
For example, âcM12â for âcentrifugal medulla-targeting 12â.
Note that new names were also given to non-canonical, generic hemibrain types, such as IB006. All new names are recorded in the cell_type column in the Supplementary Data.
The majority of VPNs (99.6%) and VCNs (98.3%) were assigned to specific types. Only 29 VPNs and 9 VCNs could not be confidently assigned a cell type and were therefore left untyped.
Sensory and motor neurons
We identified all non-visual sensory and motor neurons entering/exiting the brain through the antennal, eye, occipital and labial nerves by screening all axon profiles in a given nerve.
Sensory neurons were further cross-referenced to existing literature to assign modalities (through the class field) and, where applicable, a cell type. Previous studies have identified almost all head mechanosensory bristle and taste peg mechanosensory neurons102 in the left hemisphere (at the time of publication: right hemisphere). Gustatory sensory neurons were previously identified in ref. 103 and Johnstonâs organ neurons in refs. 104,105 in a version of the FAFB that used manual reconstruction (https://fafb.catmaid.virtualflybrain.org). Those neurons were identified in the FlyWire instance by transformation and overlay onto FlyWire space as described previously102.
Johnstonâs organ neurons in the right hemisphere were characterized based on innervation of the major AMMC zones (A, B, C, D, E and F), but not further classified into subzone innervation as shown previously104. Other sensory neurons (mechanosensory bristle neurons, taste peg mechanosensory neurons and gustatory sensory neurons) in the right hemisphere were identified through NBLAST-based matching of their mirrored morphology to the left hemisphere and expert review. Olfactory, thermosensory and hygrosensory neurons of the antennal lobes were identified through their connectivity to cognate uniglomerular projection neurons and NBLAST-based matching to previously identified hemibrain neurons40,106.
Visual sensory neurons (R1â6, R7â8 and ocellar photoreceptor neurons) were identified by manually screening neurons with pre-synapse in either the lamina, the medulla and/or the ocellar ganglia93.
ANs and DNs
We seeded all profiles in a cross-section in the ventral posterior GNG through the cervical connective to identify all neurons entering and exiting the brain at the neck. We identified all DNs based on the following criteria: (1) soma located within the brain dataset; and (2) main axon branch leaving the brain through the cervical connective.
We next classified the DNs based on their soma location according to a previous report107. In brief, the soma of DNa, DNb, DNc and DNd is located in the anterior half (a, anterior dorsal; b, anterior ventral; c, in the pars intercerebralis; d, outside cell cluster on the surface) and DNp in the posterior half of the central brain. DNg somas are located in the GNG.
To identify DNs described in ref. 107 in the EM dataset, we transformed the volume renderings of DN GAL4 lines into FlyWire space. Displaying EM and LM neurons in the same space enabled accurate matching of closely morphologically related neurons. For DNs without available volume renderings, we identified candidate EM matches by eye, transformed them into JRC2018U space and overlaid them onto the GAL4 or Split GAL4 line stacks (named in ref. 107 for that type) in FIJI for verification. Using these methods, we identified all but two (DNd01 and DNg25) in FAFB/FlyWire and annotated their cell type with the published nomenclature. All other unmatched DNs received a systematic cell type consisting of their soma location, an âeâ for EM type and a three digit number (for example, DNae001). A detailed account and analysis of DNs has been published108 separately.
ANs were identified based on the following criteria: (1) no soma in the brain; and (2) main branch entering through the neck connective (note that some ANs make a dendrite after entry through the neck connective and then an axon).
To distinguish sensory ascending (SA) neurons from ANs, we analysed SA neuron morphology in the male VNC dataset MANC109,110. First, we identified which longitudinal tract they travel to ascend to the brain111 and then found GAL4 lines matching their VNC morphology. We next identified putative matching axons in the brain dataset by morphology and tract membership. A detailed description of this process and the lines used has been published separately108.
FAFB laterality
In the fly brain, the asymmetric body is reproducibly around 4 times larger on the right hemisphere than on the left112,113,114, except in rare cases of situs inversus114,115. However, completion of the FlyWire whole-brain connectome and associated cell typing showed the asymmetric body to be larger on the apparent left side of the brain rather than the right, suggesting an inversion of the leftâright axis during initial acquisition of EM images comprising the FAFB dataset17. This hypothesis was confirmed by comparing of FAFB sample grids imaged using differential interference contrast microscopy to low-magnification views of corresponding EM image mosaics using CATMAID or neuroglancer. Grids were chosen with particularly obvious staining and sample preparation artefacts visible both in the differential interference contrast and low-magnification EM images (Extended Data Fig. 1), confirming that a leftâright axis inversion had taken place during image acquisition.
Owing to the extensive post-processing of the FAFB dataset and derived datasets (for example, transformation fields, image mosaicing and stack registrations to produce aligned volumes, segmentation supervoxels, proofread neuron segmentations, skeletons, meshes and myriad 3D visualizations), which had been undertaken at the time at which this error was discovered, we deemed it impractical to correct this error at the raw data level. Instead, we break a convention of presentation: usually, frontal views of the fly brain place the flyâs right on the viewerâs left. Instead, in this paper, frontal views of the fly brain place the flyâs right on the viewerâs rightâsimilar to the view one has of oneself while looking in a mirror. This maintains consistency with past publications. However, note that all labels of left and right in the figures in this paper, our companion papers, the supplemental annotations and associated digital repositories (for example, https://codex.flywire.ai, FAFB/FlyWire CATMAID) have been corrected to reflect the error during data acquisition. In these resources, a neuron labelled as being on the left is indeed on the left of the flyâs brain.
For consistency with visualizations and datasets obeying the standard convention (flyâs right on viewerâs left), FlyWire data can be mirrored. To facilitate this, we provide tools to digitally mirror FAFB-FlyWire data using the Python flybrains (https://github.com/navis-org/navis-flybrains) or natverse nat.jrcbrains (https://github.com/natverse/nat.jrcbrains) packages (Extended Data Fig. 1c), through the
navis.mirror_brain()
and
nat.jrcbrains::mirror_fafb()
function calls, respectively. See the fafbseg-py documentation for a tutorial on mirroring.
We also provide a neuroglancer scene in which both FlyWire and hemibrain data are displayed in the correct orientation: https://tinyurl.com/flywirehbflip783. In this scene, a frontal view has both FAFB and hemibrain RHS to the left of the screen, obeying the standard convention. The scene displays the SA1 and SA2 neurons, which target the right asymmetric body for both FlyWire and the hemibrain, confirming that the RHS for both datasets has been superimposed (compare with Extended Data Fig. 1a).
Morphological comparisons
Throughout our analyses, NBLAST38 was used to generate morphological similarity scores between neuronsâfor example, for matching neurons between the FlyWire and the hemibrain datasets, or for the morphological clustering of the hemilineages. In brief, NBLAST treats neurons as point clouds with associated tangent vectors describing directionality, so called dotprops. For a given queryâtarget neuron pair, we perform a k-nearest neighbours search between the two point clouds and score each nearest-neighbour pair by their distance and the dot product of their vector. These are then summed up to compute the final queryâtarget NBLAST score. It is important to note that direction of the NBLAST matters, that is, NBLASTing neurons AâBâ BâA. Unless otherwise noted, we use the minimum between the forward and reverse NBLAST scores.
The NBLAST algorithm is implemented in both navis and the natverse (Table 1). However, we modified the navis implementation for more efficient parallel computation in order to scale to pools of more than 100,000 neurons. For example, the all-by-all NBLAST matrix for the full 139,000 FlyWire neurons alone occupies over 500âGB of memory (32âbit floats). Most of the large NBLASTs were run on a single cluster node with 112 CPUs and 1âTB RAM provided by the MRC LMB Scientific Computing group, and took between 1 and 2 days (wall time) to complete.
Below, we provide recipes for the different NBLAST analyses used in this paper:
FlyWire all-by-all NBLAST
For this NBLAST, we first generated skeletons using the L2 cache. In brief, underlying the FlyWire segmentation is an octree data structure where level 0 represents supervoxels, which are then agglomerated over higher levels116. The second layer (L2) in this octree represents neurons as chunks of roughly 4âÃâ4âÃâ10âμm in size, which is sufficiently detailed for NBLAST. The L2 cache holds precomputed information for each L2 chunk, including a representative x/y/z coordinate in space. We used the x/y/z coordinates and connectivity between chunks to generate skeletons for all FlyWire neurons (implemented in fafbseg; Table 1). Skeletons were then pruned to remove side branches smaller than 5âμm. From those skeletons, we generated the dotprops for NBLAST using navis.
Before the NBLAST, we additionally transformed dotprops to the same side by mirroring those from neurons with side right onto the left. The NBLAST was then run only in forward direction (queryâtarget) but, because the resulting matrix was symmetrical, we could generate minimum NBLAST scores using the transposed matrix: min(Aâ+âAT).
This NBLAST was used to find leftâright neuron pairs, define (hemi)lineages and run the morphology group clustering.
FlyWireâhemibrain NBLAST
For FlyWire, we re-used the dotprops generated for the all-by-all NBLAST (see the previous section). To account for the truncation of neurons in the hemibrain volume, we removed points that fell outside the hemibrain bounding box.
For the hemibrain, we downloaded skeletons for all neurons from neuPrint (https://neuprint.janelia.org) using neuprint-python and navis (Table 1). In addition to the approximately 23,000 typed neurons, we also included all untyped neurons (often just fragments) for a total of 98,000 skeletons. These skeletons were pruned to remove twigs smaller than 5âμm and then transformed from hemibrain into FlyWire (FAFB14.1) space using a combination of non-rigid transforms116,117 (implemented through navis, navis-flybrain and fafbseg; Table 1). Once in FlyWire space, they were resampled to 0.5 nodes per μm of cable to approximately match the resolution of the FlyWire L2 skeletons, and then turned into dotprops. The NBLAST was then run both in forward (FlyWire to hemibrain) and reverse (hemibrain to FlyWire) direction and the minimum between both were used.
This NBLAST allowed us to match FlyWire left against the hemibrain neurons. To also allow matching FlyWire right against the hemibrain, we performed a second run after mirroring the FlyWire dotprops to the opposite side.
In Fig. 3c,d, we manually reviewed NBLAST matches. For this, we sorted hemibrain neurons based on their highest NBLAST score to a FlyWire neuron into bins with a width of 0.1. From each bin, we picked 30 random hemibrain neurons (except for bin 0â0.1 which contained only 27 neurons in total) and scored their top five FlyWire matches as to whether a plausible match was among them. In total, this sample contained 237 neurons.
Cross-brain co-clustering
The pipeline for the morphology-based across brain co-clustering used in Fig. 6 and Extended Data Fig. 9 was essentially the same as for the FlyWireâhemibrain NBLAST with two exceptions: (1) we used high-resolution FlyWire skeletons instead of the coarser L2 skeletons (see below); and (2) both FlyWire and hemibrain skeletons were resampled to 1 node per μm before generating dotprops.
High-resolution skeletonization
In addition to the coarse L2 skeletons, we also generated high-resolution skeletons that were, for example, used to calculate the total length of neuronal cable reported in our companion paper1 (149.2âm). In brief, we downloaded neuron meshes (LOD 1) from the flat 783 segmentation (available at gs://flywire_v141_m783) and skeletonized them using the wavefront method implemented in skeletor (https://github.com/navis-org/skeletor). Skeletons were then rerooted to their soma (if applicable), smoothed (by removing small artifactual bristles on the backbone), healed (segmentation issues can cause breaks in the meshes) and slightly downsampled. A modified version of this pipeline is implemented in fafbseg. Skeletons are available for download (see the âData availabilityâ and âCode availabilityâ sections).
Connectivity normalization
Throughout this paper, the basic measure of connection strength is the number of unitary synapses between two or more neurons79; connections between adult fly neurons can reach thousands of such unitary synapses2. Previous work in larval Drosophila has indicated that synaptic counts approximate contact area118, which is most commonly used in mammalian species when a high-resolution measure of anatomical connection strength is required. Connectomics studies also routinely use connection strength normalized to the target cellâs total inputs71,79. For example, if neurons i and j are connected by 10 synapses and neuron j receives 200 inputs in total, the normalized connection weight i to j would be 5%. A previous study119 showed that while absolute number of synapses for a given connection changes drastically over the course of larval stages, the proportional (that is, normalized) input to the downstream neuron remains relatively constant119. Importantly, we have some evidence (Fig. 4g) that normalized connection weights are robust against technical noise (differences in reconstruction status, synapse detection). Note that, for analyses of mushroom body circuits, we use an approach based on the fraction of the input or output synaptic budget associated with different KC cell types; this differs slightly from the above definition and will be detailed in a separate section below.
Connectivity stereotypy analyses
For analyses on connectivity stereotypy (Fig. 4 and Extended Data Fig. 6) we excluded a number of cell types:
-
KCs, due to the high variability in numbers and synapse densities in the mushroom body lobes between FlyWire and the hemibrain (Fig. 5 and Extended Data Fig. 7).
-
Cell types that exist only on the left but not the right hemisphere of the hemibrain because our comparison was principally against the right hemisphere.
-
Antennal lobe receptor neurons, because truncation/fragmentation in the hemibrain causes some ambiguity with respect to their side annotation.
-
Cell types with members that have been marked as being affected by sample or imaging artefacts (that is, status âoutlier_segâ).
-
VPNs, as they are heavily truncated in the hemibrain.
Among the remaining types, we used only the 1:1 and 1:many but not the many:1 matches. Taken together, we used 2,954 (hemibrain) types for the connectivity stereotypy analyses.
Availability through CATMAID Spaces
To increase the accessibility and reach of the annotated FlyWire connectome, meshes of proofread FlyWire neurons and synapses were skeletonized and imported into CATMAID, a widely used web-based tool for collaborative tracing, annotation and analysis of large-scale neuronal anatomy datasets79,120 (https://catmaid.org; Extended Data Fig. 10). Spatial annotations like skeletons are modelled using PostGIS data types, a PostgreSQL extension that is popular in the geographic information system community. This enables us to reuse many existing tools to work with large spatial datasets, for example, indexes, spatial queries and mesh representation.
A publicly available version of the FlyWire CATMAID project is available online (https://fafb-flywire.catmaid.org). This project uses a new extension, called CATMAID Spaces (https://catmaid.org/en/latest/spaces.html), which allows users to create and administer their own tracing and annotation environments on top of publicly available neuronal image volumes and connectomic datasets. Moreover, users can now login through the public authentication service ORCiD (https://www.orcid.org), so that everyone can log-in on public CATMAID projects. Users can also now create personal copies (Spaces) of public projects. The user then becomes an administrator, and can invite other users, along with the management of their permissions in this new project. Invitations are managed through project tokens, which the administrator can generate and send to invitees for access to the project. Both CATMAID platforms can talk to each other and it is possible to load data from the dedicated FAFB-FlyWire server in the more general Spaces environment.
Metadata annotations for each neuron (root id, cell type, hemilineage, neurotransmitter) were imported for FlyWire project release 783. Skeletons for all 139,255 proofread neurons were generated from the volumetric meshes (see the âHigh-resolution skeletonizationâ section) and imported into CATMAID, resulting in 726,831,877 treenodes. To reduce the import time, skeletons were imported into CATMAID directly as database inserts through SQL, rather than through public RESTful APIs. FlyWire root IDs are available as metadata for each neuron, facilitating interchange with related resources such as FlyWire Codex1. Synapses attached to reconstructed neurons were imported as CATMAID connector objects and attached to neuron skeletons by doing a PostgreSQL query to find the nearest node on each of the partner skeletons. Connector objects were linked to postsynaptic partners only if the downstream neuron was in the proofread data release (180,016,288 connections from the 130,054,535 synapses with at least one partner in the proofread set).
Synapse counts
Insect synapses are polyadic, that is, each presynaptic site can be associated with multiple postsynaptic sites. In contrast to the Janelia hemibrain dataset, the synapse predictions used in FlyWire do not have a concept of a unitary presynaptic site associated with a T-bar46. Thus, pre-synapse counts used in this paper do not represent the number of presynaptic sites but rather the number of outgoing connections.
In Drosophila connectomes, reported counts of the inputs (post-synapses) onto a given neuron are typically lower than the true number. This is because fine-calibre dendritic fragments frequently cannot be joined onto the rest of the neuron, instead remaining as free-floating fragments in the dataset.
Technical noise model
To model the impact of technical noise such as proofreading status and synapse detection on connectivity, we first generated a fictive â100%â ground-truth connectivity. We took the connectivity between cell-typed left FlyWire neurons and scaled each edge weight (the number of synapses) by the postsynaptic completion rates in the respective neuropil. For example, all edge weights in the left mushroom body calyx (CA), which has a postsynaptic completion rate of 52.5%, were scaled by a factor of 100/52.5â=â1.9.
In the second step, we simulated the proofreading process by randomly drawing (without replacement) individual synaptic connections from the fictive ground-truth until reaching a target completion rate. We further simulate the impact of false positives and false negatives by randomly adding and removing synapses to/from the draw according to the precision (0.72) and recall (0.77) rates reported previously46. In each round, we made two draws: (1) A draw using the original per-neuropil postsynaptic completion rates; and (2) a draw where we flip the completion rates for left and right neuropils, that is, use the left CA completion rate for the right CA and vice versa.
In each of the 500 rounds that we ran, we drew two weights for each edge. Both stem from the same fictive 100% ground-truth connectivity but have been drawn according to the differences in left versus right hemisphere completion rates. Combining these values, we calculated the mean difference and quantiles as function of the weight for the FlyWire left (that is, the draw that was not flipped) (Fig. 4i). We focussed this analysis on edge weights between 1 and 30 synapses because the frequency of edges stronger than that is comparatively low, leaving gaps in the data.
KC analyses
Connection weight normalization and synaptic budget analysis
When normalizing connection weights, we typically convert them to the percentage of total input onto a given target cell (or cell type). However, in the case of the mushroom body, the situation is complicated by what we think is a technical bias in the synapse detection methods used for the two connectomes that causes certain kinds of unusual connections to be very different in frequency between the two datasets. We find that the total number of post-synapses as well as the post-synapse density in the mushroom body lobes are more than doubled in the hemibrain compared with in FlyWire (Extended Data Fig. 7b,c). This appears to be explained by certain connections (especially KC to KC connections, which are predominantly arranged with an unusual rosette configuration along axons and of which the functional significance is poorly understood121) being much more prevalent in the hemibrain than in FlyWire (Extended Data Fig. 7d). Some other neurons, including the APL giant interneuron, also make about twice as many synapses onto KCs in the hemibrain compared with in FlyWire (Extended Data Fig. 7a). As a consequence of this large number of inputs onto KC axons in the hemibrain, input percentages from all other cells are reduced in comparison with FlyWire.
To avoid this bias, and because our main goal in the KC analysis was to compare different populations of KCs, we instead expressed connectivity as a fraction of the total synaptic budget for upstream or downstream cell types. For example, we examined the fraction of the APL output that is spent on each of the different KC types. Similarly, we quantified connectivity for individual KCs as a fraction of the budget for the whole KC population.
Calculating K from observed connectivity
Calculation of K, that is, the number of unique odour channels that each KC receives input from, was principally based on their synaptic connectivity. For this, we looked at their inputs from uniglomerular ALPNs and examined from how many of the 58 antennal lobe glomeruli does a KC receive input from. K as reported in Fig. 6 is based on non-thresholded connectivity. Filtering out weak connections does lower K but, importantly, our observations (for example, that KCg-m cells have a lower K in FlyWire than in the hemibrain) are stable across thresholds (Extended Data Fig. 7g).
KC model
A simple rate model of neural networks122 was used to generate the theoretical predictions of K, the number of ALPN inputs that each KC receives (Fig. 5k). KC activity is modelled by
$${\bf{h}}={\bf{W}}\cdot {{\bf{r}}}_{{\rm{P}}{\rm{N}}},$$
where h is a vector of length M representing KC activity, \({\bf{W}}\) is an MâÃâN matrix representing the synaptic weights between the KCs and PNs, rPN is a vector of length N representing PN activity. The number of KCs and ALPNs is denoted by M and N, respectively. In this model, the PN activity is assumed to have zero mean, \({\bar{{\bf{r}}}}_{{\rm{P}}{\rm{N}}}=0\), and be uncorrelated, \(\bar{{{\bf{r}}}_{{\rm{P}}{\rm{N}}}\cdot {{\bf{r}}}_{{\rm{P}}{\rm{N}}}}={{\bf{I}}}_{N}\). Here, \({{\bf{I}}}_{N}\) is an NâÃâN identity matrix and \({\bar{{\bf{r}}}}_{{\rm{P}}{\rm{N}}}\) denotes the average taken over independent realizations of \({{\bf{r}}}_{{\rm{P}}{\rm{N}}}\). Then, the ijth element of the covariance matrix of h is
$$[{\bf{C}}{]}_{ij}=\bar{{[{\bf{h}}]}_{i}{[{\bf{h}}]}_{j}}=\mathop{\sum }\limits_{k=0}^{N}[{\bf{W}}{]}_{ik}{[{\bf{W}}]}_{jk}.$$
More detailed calculations can be found in a previous report122. Randomized and homogeneous weights were used to populate \({\bf{W}}\), such that each row in \({\bf{W}}\) has K elements that are 1âââα and NâââK elements that are âα. The parameter α represents a homogeneous inhibition corresponding to the biological, global inhibition by APL. The value inhibition was set to be αâ=âA/M, where Aâ=â100 is an arbitrary constant and M is the number of KCs in each of the three datasets. The primary quantity of interest is the dimension of the KC activities defined by122:
$$\dim ({\bf{h}})=\frac{{(\text{Tr}[{\bf{C}}])}^{2}}{\text{Tr}[{{\bf{C}}}^{2}]}$$
and how it changes with respect to K, the number of input connections. In other words, what are the numbers of input connections K onto individual KCs that maximize the dimensionality of their responses, h, given M KCs, N ALPNs and a global inhibition α?
From Fig. 5k, the theoretical values of K that maximize dim(h) in this simple model demonstrate the consistent shift towards lower values of K found in the FlyWire left and FlyWire right datasets when compared with the hemibrain.
The limitations of the model are as follows:
-
(1)
The values in the connectivity matrix \({\bf{W}}\) take only two discrete values, either 0 and 1 or 1âââα and α. In a way, this helps when calculating analytical results for the dimensionality of the KC activities. However, it is unrealistic as the connectomics data give the number of synaptic connections between the ALPNs and the KCs.
-
(2)
The global inhibition provided by APL to all of the mixing layer neurons is assumed to take a single value for all neurons. In reality, the level of inhibition would be different depending on the number of synapses between APL and the mixing layer neurons.
-
(3)
It is unclear whether the simple linear rate model presented in the original paper represents the behaviour of the biological neural circuit well. Furthermore, it remains unproven that the ALPN-KC neural circuit is attempting to maximize the dimensionality of the KC activities, albeit the theory is biologically well motivated (but see refs. 49,50).
-
(4)
The number of input connections to each mixing layer neuron is kept at a constant K for all neurons. It is definitely a simplification that can be corrected by introducing a distribution P(K) but this requires further detailed modelling.
Statistical analyses
Unless otherwise stated, statistical analyses (such as Pearson R or cosine distance) were performed using the implementations in the scipy123 Python package. To determine statistical significance, we used either t-tests for normally distributed samples, or KolmogorovâSmirnov tests otherwise.
Cohenâs d124 was calculated as follows:
$$d=\frac{{\bar{x}}_{1}-{\bar{x}}_{2}}{s}$$
where pooled s.d. s is defined as:
$$s=\sqrt{\frac{({n}_{1}\,-\,1){s}_{1}^{2}\,+\,({n}_{2}\,-\,1){s}_{2}^{2}}{{n}_{1}\,+\,{n}_{2}\,-\,2}}$$
where the variance for one of the groups is defined as:
$${s}_{1}^{2}=\frac{1}{{n}_{1}-1}{\sum }_{i=1}^{{n}_{1}}{({x}_{1,i}-{\bar{x}}_{1})}^{2}$$
and similar for the other group.
Enhanced box plotsâalso called letter-value plots125âin Fig. 5h and Extended Data Fig. 7f are a variation of box plots better suited to represent large samples. They replace the whiskers with a variable number of letter values where the number of letters is based on the uncertainty associated with each estimate, and therefore on the number of observations. The âfattestâ letters are the (approximate) 25th and 75th quantiles, respectively, the second fattest letters the (approximate) 12.5th and 87.5th quantiles and so on. Note that the width of the letters is not related to the underlying data.
Mapping to the VirtualFlyBrain database
The VirtualFlyBrain (VFB) database22 curates and extracts information from all publications relating to Drosophila neurobiology, especially neuroanatomy. The majority of published neuron reconstructions, including those from the hemibrain, can be examined in the VFB. Each individual neuron (that is, one neuron from one brain) has a persistent ID (of the form VFB_xxxxxxxx). Where cell types have been defined, they have an ontology ID (for example, FBbt_00047573, the ID for the DNa02 DN cell type). Importantly, VFB cross-references neuronal cell types across publications even if different terms were used. It also identifies driver lines to label many neurons. In this paper, we generate an initial mapping providing FBbt IDs for the closest and fine-grained ontology term that already exists in their database. For example, a FlyWire neuron with a confirmed hemibrain cell type will receive a FBbt ID that maps to that exact cell type, while a DN that has been given a new cell type might only map to the coarser term âadult descending neuronâ. Work is already underway with the VFB to assign both ontology IDs (FBbt) to all FlyWire cell types as well as persistent VFB_ids to all individual FlyWire neurons.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.