Overview
Our study analysed the FAFB, an adult female D. melanogaster brain imaged at the synaptic level resolution with serial section transmission electron microscopy17. We used the FlyWire interface, which auto-segmented FAFB EM data to construct three-dimensional segmentations of individual neurons29. To reconstruct desired neurons, we first identified relevant axons, dendrites and branches. Possible errors by the auto-segmentation were mainly unfinished branches caused by missing EM slices or incorrect connections caused by shifted EM slices. In addition, some neurons had darker cytosols in the EM data, possibly owing to neuronal damage during the dissection process61, and were therefore not as well-constructed by the auto-segmentation. We manually corrected each of these errors.
Dense EM reconstruction
To find all MeTu, TuBu, TuTu and AOTU046 neurons in the AVP, we densely reconstructed the AOTUsu by scanning through every layer of EM in the neuropil volumes and proofreading all neurons composing them (disregarding twigs). ER neurons were identified by following TuBu downstream connectivity30. After each of these neurons was proofread, we classified them and compiled lists of their coordinates for further analysis. See Supplementary Table 1 for details of the editing and naming history.
Region boundaries
Regions were distinguished in our study so as to limit synapses to specified neuropils. These regions included ME_L, ME_R, LO_L, LO_R, AOTU_L, AOTU_R, BU_L, BU_R and EB (in which ME indicates medulla; BU, bulb; LO, lobula; L, left; R, right; and EB, ellipsoid body). Using SciPyâs spatial module, we created Delaunay tessellations using a set of FlyWire coordinates to determine whether synapses were contained within the given regions. The sets of points were not a comprehensive boundary box of individual neuropils, but rather formed polyhedra that contained the ROIs of the relevant neurons. The coordinates were selected with the help of FlyWireâs annotation lines to ensure that all neuronsâ synapses were incorporated. In the case of the medullas, in which the Delaunay tessellation incorporated some lobula synapses as well, the ipsilateral lobula region was subtracted.
AOTUsu subdivision in comparison with previous studies
The connectome of the AVP, which revealed four major MeTu types, clarifies discrepancies in previous literature. MeTuim in Omoto & KeleŠet al.39 seems to be MeTu4 because DALcl2d TuBu neurons project from AOTUim to the inferior bulb (BUi). In addition, on the basis of spatial organization, AOTUlc, AOTUlp and AOTUla/il in Omoto & KeleŠet al. might correspond to MeTu1, MeTu2 and MeTu3 locations, respectively, although the AOTUsu map is slightly different from our study (Extended Data Fig. 1d). A crucial discrepancy we could not resolve was TuBua in Omoto & KeleŠet al. They described that TuBua projects from AOTUil to the anterior BU (BUa), which we did not observe in the FAFB dataset. In Hulse, Haberkern, Franconville and Turner-Evans et al.16, the TuBu neuron type innervating the BUa is TuBu01, which are located in AOTUsu_PC, downstream of MeTu2. However, Omoto & Keles et al. say that these TuBu neurons project from the AOTUsu_A to the BUa. We believe that this discrepancy is due to the lower resolution of light microscopy, and think that TuBua should be reclassified.
Timaeus et al.14 divided the AOTUsu into five subdomains, separating the AOTUsu_A into lateral and anterior central parts. They state that R7 might be upstream of MeTula, MeTuca and MeTucp (MeTu3 and MeTu2, respectively), which agrees with what we found. However, they only found TuBu neurons projecting from the AOTUsula, AOTUsulp, AOTUsuca or AOTUsucp to the BUs and the AOTUsum to the BUi, meaning that they did not discover TuBu01. Finally, they found that MeTum (MeTu4) dendrites also projected to medulla layers 2 and 8, which was inconsistent with what we found in the FAFB dataset.
Tai et al.15, unlike the other two papers, found four subdomains of the AOTUsu (L-AOTU1â4), which are connected linearly from the edge of the AOTUlu to the lateral-most edge of the AOTUsu. The respective MeTu neurons in these regions were called MT1â4 (not corresponding to our studyâs MeTu1âMeTu4). This study only showed an anterior view of the AOTU, and as such, it is possible that they did not find the AOTUsu_PC, which is obscured by the AOTUsu_A from the anterior side. In this case, the corresponding regions are AOTUsu_M (L-AOTU1), AOTUsu_A (L-AOTU2â3) and AOTUsc_PL (L-AOTU4). The corresponding MeTu neurons are thus MeTu4a, MeTu4b, MeTu4c and MeTu4d (MT1), MeTu3c (MT2), MeTu3a and MeTu3b (MT3) and MeTu1 (MT4).
Synaptic connectivity matrices
Synaptic connectivity between neurons was found using automatic synapse detection30. For all our connectivity analyses, we used a cleft score of â¥50 and excluded autapses and synapses to the background segmentation. Two types of connectivity matrices were generated throughout the study: Supplementary Data 1aâg show individual neuron weight matrices (purple) and neural type weight matrices (green). For the individual neuron weight matrices (Supplementary Data 1aâd), the number of synapses between each neuron was first calculated. To determine the relative weight within the given region, this quantity was divided by the postsynaptic neuronâs total number of synapses in the region.
Certain outlier neurons heavily skewed the colour plot matrices because they had few connections in their respective regions or nearly exclusively received synaptic weight from a single neuron. To resolve the former issue, neurons with fewer than five total regional connections were not included in the matrices. To resolve the latter issue, a small number of outliers were removed from medulla MeTu interconnectivity plots: one MeTu1_R, one MeTu2a_R and three MeTu4a_R.
The ordering of the neurons within the connectivity matrices was based on the location of TuBu neurons along the dorsalâventral axis within the AOTUsu. Both MeTu and ER neurons were ordered in groups according to which of these TuBu neurons they were most connected to (MeTu neurons presynaptically in the AOTUsu and ER neurons postsynaptically in the bulb). Within the groups they were ordered by how many synapses they shared with that TuBu neuron.
Neural type weight matrices (Supplementary Data 1eâg) show the connections of whole classes of neurons. First, the total number of synapses between all presynaptic and postsynaptic neurons of the respective given types was calculated. Then, these quantities were divided by the total number of synapses of all postsynaptic neurons of the given type within the region. This gave a measure of the total synaptic weight between the two types.
Three-dimensional rendering
Three-dimensional renderings were either generated in Blender with neuron meshes retrieved using the Python CloudVolume package or in R with the rgl and fafbseg package.
Medulla columns and layers
We identified all Mi1 neurons, a unicolumnar cell type, in both hemispheres as a proxy for individual medulla columns because Mi1 neurons are present in each medulla column and span the entire distalâproximal axis of the medulla from layer M1 to layer M10. For each Mi1 neuron we performed a principal component analysis (PCA) on all pre- and postsynaptic sites of the neuron (Extended Data Fig. 1e). PC1 corresponds to the distalâproximal axis of the column. The upper and lower boundary of each column is defined as the 0.03 and 0.97 percentile of synapses on the distalâproximal axis.
Medulla layers are based on the average synapse distribution of Mi1, Mi4, L1, L2, L3, L5, Dm8 and T4 neurons along the distalâproximal axis in three exemplar columns. Layer M1: [â3.9â5.5%]; layer M2: [5.5â17.1%]; layer M3: [17.1â30.8%]; layer M4: [30.8â34.0%]; layer M5: [34.0â43.2%]; layer M6: [43.2â50.1%]; layer M7: [50.1â63.1%]; layer M8: [63.1â75.4%]; layer M9: [75.4â92.4%]; layer M10: [92.4â102.2%].
MeTu classification
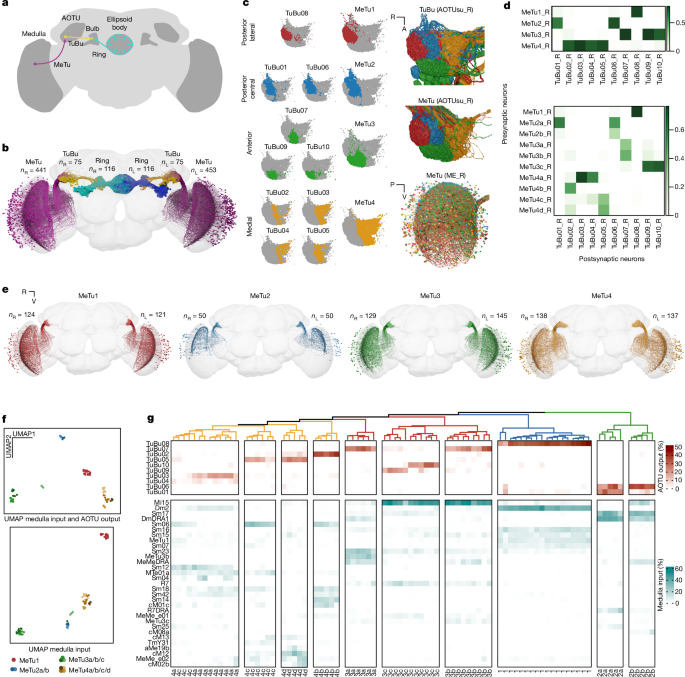
We describe MeTu types (labelled with numbers: MeTu1âMeTu4) and MeTu subtypes (labelled with lowercase letters; for example, MeTu2a). MeTu1, MeTu2 and MeTu3 were previously called MC61 (ref. 62) and MeTu4 was called MC64 (ref. 16). MeTu2 was also called MeTu-DRA37. The location of axons and dendrites of MeTu (Fig. 1c), TuBu (Fig. 1c), TuTu (Extended Data Fig. 4) and AOTU046 (Extended Data Fig. 4) neurons maintain specific patterns of processes within the AOTUsu16, through which we determined four distinct regions (posterior lateral, posterior central, anterior and medial). The axonal boutons of each MeTu neuron terminate within one of these four areas, so we classified MeTu1, MeTu2, MeTu3 and MeTu4 as types. Between the left and right hemispheres, respectively, there are 121 and 124 MeTu1, 50 and 50 MeTu2, 145 and 129 MeTu3 and 137 and 138 MeTu4. There was one neuron on the right side whose axonal tract terminated before projecting to the medulla. It was labelled MeTu_incomplete_R and was excluded from further analysis.
Analysis of morphology, up- and downstream connectivity as well as spatial distribution in the medulla revealed distinct MeTu subpopulations within MeTu2, MeTu3 and MeTu4, which led us to define MeTu subtypes.
MeTu1 forms a homogenous neuron population in terms of morphology, and up- and downstream connectivity, without any distinctive features that would allow any further subtyping (Fig. 2m). MeTu2a is connected to both TuBu01 and TuBu06 with a preference for TuBu01, while MeTu2b is primarily connected to TuBu06 with very few synapses onto TuBu01 (Extended Data Fig. 5k).
We found three MeTu3 subtypes: MeTu3a, MeTu3b and MeTu3c. MeTu3a has flat dendrites and lacks presynaptic connections to Mi15, whereas MeTu3b and MeTu3c have vertical dendritic protrusions and connect to Mi15Â (Extended Data Fig. 5d,e). MeTu3a was specifically classified as MeTu3 that has 13 or fewer synapses with Mi15 neurons. Note that MeTu2a and MeTu2b cell bodies are located closer to the medulla equator, whereas MeTu3a cell bodies are found above the centre of the branching (data not shown). Within the AOTUsu, all MeTu3a project to TuBu07. MeTu3b is strongly connected to TuBu07, and MeTu3c is most strongly connected to TuBu09 and TuBu10. To further analyse this distinction, we compared their postsynaptic weights with Mi15, Sm17, Sm23 and MeMeDRA. Some MeTu cells sensitive to skylight polarization have so far been physiologically characterized in Drosophila11, and a careful comparison between their light microscopic data and our connectomic reconstruction identifies these cells as MeTu2b and MeTu3a. Finally, MeTu3c might have subpopulations: MeTu3c_dorsal and MeTu3c_ventral, on the basis of the TuBu connectivity. Their connectivity in the medulla was indistinguishable other than the general location (dorsal medulla versus ventral medulla), which might suggest the same kind of information processing. Furthermore, the axons to downstream TuBu09 and TuBu10 overlap somewhat, suggesting that functional differences may occur downstream ofâbut not atâthe MeTu3c neurons. For these reasons, we decided to combine them into a single subtype.
MeTu4 is generally morphologically distinct from other MeTu types because neurons contain boutons within the lobula. However, light microscopy suggested there is a subtype that does not have these boutons (Extended Data Fig. 8h). We also found a MeTu4 population without lobula boutons and few lobula synapses (fewer than 15 pre- and postsynapses), which we named MeTu4d. MeTu4d in addition only arborizes within the ventral half of the medulla.
We further grouped MeTu4 neurons with lobula boutons into distinctive subtypes on the basis of downstream TuBu connectivity. MeTu4a are presynaptic to TuBu03 and TuBu04, MeTu4b are presynaptic to TuBu02 and MeTu4c are presynaptic to TuBu05.
UMAP in Fig. 1f (top) is based on connectivity to up- and downstream partners as features. We selected a total of 84 neurons (see âUpstream connectionsâ for more information). Downstream neurons include 13 types (all TuBu types, TuTuA, TuTuB and AOTU046), and upstream neurons include 28 types (all top 5 connected neuron types of all MeTu subtypes. UMAP in Fig. 1fii is based only on the 31 upstream types (28 non-MeTu and 3 MeTu types). All connectivity types are also shown in Fig. 1g.
As the entire optic lobe connectivity became available (FlyWire v.783)45, we also performed the same analysis using the entire dataset in the right hemisphere (Supplementary Data 1h).
Finally, we sought to provide light microscopic evidence in the form of cell-type-specific driver lines, corroborating the existence of the genetically defined subclasses of visual projection neurons that are described in this study33,34,63,64 (see Supplementary Table 2).
Proofreading rounds
For a subset of MeTu neurons described in the previous section, we increased the proofreading quality by increasing the rounds of detailed proofreading29. We used the right optic lobe because the left optic lobe has a partially detached lamina and parts of the posterior side of the medulla are distorted17. We chose 113 of the 441 right MeTu neurons to undergo multiple rounds of proofreading. Originally, 101 neurons were chosen randomly with the same relative ratios of MeTu1âMeTu4 neurons as in the population: 28 MeTu1, 12 MeTu2, 30 MeTu3 and 31 MeTu4. When we later discovered subcategories of the neurons, we wanted at least five of each subtype. In the end, we proofread the following 113 neurons: 29 MeTu1, 7 MeTu2a, 5 MeTu2b, 6 MeTu3a, 13 MeTu3b, 16 MeTu3c, 13 MeTu4a, 5 MeTu4b, 14 MeTu4c and 5 MeTu4d.
Each of these neurons underwent three rounds of proofreading, and synaptic and skeletal comparisons were performed to determine the differences in accuracy between the three rounds. The first round was the cursory proofreading that was done to all 441 MeTu neurons. The next two rounds were split between the two proofreaders (D.G. and E.K.). Each proofreader densely proofread half of the 113 for the second round, and then switched and worked on the other half for the third round. Afterwards, F1 scores were computed on both the number of synapses and the number of skeletal nodes of each neuron between rounds. These scores showed the decrease in the number of edits between the first round and subsequent rounds.
Upstream connections
We used automatic synapse detection to find presynaptic partners of the proofread MeTu neurons. As stated in the proofreading section, we picked them on the basis of the ratio of the entire population, with a minimum of five neurons per type. In addition, as with the proofreading rounds, we only looked at neurons on the right side. Because several neurons contain a darker cytosol and are not segmented well in FlyWire, we left out any of those neurons in favour of normal neurons. Thus, we analysed the following 84 neurons: 18 MeTu1, 5 MeTu2a, 5 MeTu2b, 6 MeTu3a, 9 MeTu3b, 11 MeTu3c, 10 MeTu4a, 5 MeTu4b, 10 MeTu4c and 5 MeTu4d. For each neuron, we identified all upstream partners with five or more synapses. Many partners had been classified in previous studies, and for any others we used a nomenclature proposed recently45.
Synapse density
MeTu and TuBu synapse density maps in the AOTUsu were created from three angles: from the dorsal side looking towards the ventral side; from the anterior side looking towards the posterior side; and from the lateral side looking towards the medial side (Extended Data Fig. 3a,b). Each of these views was rotated 30° along the anteriorâposterior axis. Each map was created by finding all of the connections within small volumes, each 40ânm by 40ânm by the length of the AOTUsu along the viewpoint axis. When the number of connections was computed, they were subjected to a Gaussian blur with a sigma value of 10. Colour maps were then created on the basis of the relative values, with higher values having higher opacity. Demonstrative synapse maps were created as well (Fig. 1c). These were subjected to a Gaussian blur (with a sigma value of 4), and did not vary in opacity according to synapse density.
Neurotransmitter predictions
We used the neurotransmitter prediction described in a recent study51. We calculated the average neurotransmitter probability across all presynaptic sites of an individual neuron (Extended Data Figs. 4c,e and 12).
AVP classification
The existing connectomic analysis16 of the hemibrain65,66 provided full classifications of TuTu, TuBu and ER neurons, which we adopted in this study. This study gave detailed classifications to 17 bulbar ER neurons and 5 lateral accessory lobe ER neurons. Of the 17 bulbar neurons, there are 11 distinct morphologies, and we classified the FAFB neurons as follows: ER2_abd, ER2_c, ER3a_ad, ER3d_acd, ER3d_b, ER3m, ER3p_ab, ER3w. ER4d, ER4m and ER5. The study also described patterns of interconnectivity between ER neurons, and using synaptic analysis we distinguished ER2ad and ER2b, and ER3d_a, ER3d_c and ER3d_d. There are multiple morphologies of ER2c neurons (which is consistent with hemibrain), but we did not further subcategorize these neurons. However, some connectivity patterns are not consistent between the hemibrain and FAFB, so we did not subclassify all neurons to the same level of detail. In the instances of ER2_a and ER2_d; ER3a_a and ER3a_d; ER3p_a and ER3p_b; and ER3w_a and ER3w_b, we maintained their names as ER2_ad, ER3a_ad, ER3d_acd, ER3p_ab and ER3w_ab.
In the hemibrain, TuBu neurons were classified on the basis of their downstream ER neuron partners. After classifying all the corresponding ER neurons, we similarly grouped the TuBu neurons as TuBu01âTuBu10. There are three ambiguous TuBu neurons. One TuBu in the right hemisphere is upstream of an ER2c neuron but is located in line with other TuBu09 as opposed to TuBu10, which are generally upstream of ER2c. We labelled this neuron TuBu09 because of its location in AOTUsu. Another TuBu neuron in the right hemisphere has the dendritic morphology of a TuBu04 and is downstream of MeTu4a, but is upstream of ER3p_ab. We classified it as TuBu04 as opposed to TuBu05. One neuron in the left hemisphere has a normal microglomerulus partnered with an ER3a_ad and two ER3m neurons like TuBu02 neurons. However, this neuron projects to the SPS, as opposed to the AOTUsu. Because there is no other neuron in this dataset or hemibrain with this projection pattern, we determined that it might have been a developmental error and labelled it TuBu_misc_L, only including it in connectivity tables between TuBu and ER neurons.
We identified TuTub_a and TuTub_b on the basis of morphology. There is one of each type per hemisphere. There are four AOTU046 neurons, with dendrites in one SPS and axons in both AOTUsu and both bulbs. The quantity of each of these bihemispheric neurons is consistent with hemibrain16.
Bihemispheric connections
Connectivity diagrams of bihemispheric neurons are based on type connectivity matrices from the right hemisphere (Extended Data Fig. 4a). Each arrow represents the weight of the postsynaptic neuron typeâs connection to the presynaptic neuron type. Only weights â¥0.05 were represented as arrows. Arrow thickness was determined linearly on the basis of the weight.
The bihemispheric neuron diagrams in Extended Data Fig. 4b,d are made using neurons from the right hemisphere. Pie charts within the figures show the relative amount of presynaptic (red) and postsynaptic (cyan) connections of the neuron. Within the AOTUsu, these only include connections between the bihemispheric neurons and MeTu and TuBu neurons. Within the bulb, the connections shown are between AOTU046 and TuBu and ER neurons. Within the SPS, the connections are between AOTU046 and all SPS neurons. The relative size of the pie charts refers to the quantities of bihemispheric synapses in each subregion. In the case of AOTU046, these were calculated by averaging the two neurons on the right side. Lines are drawn to subregions that have 100 or more synapses.
Alternative visual pathways
To identify other potential visual pathways, we looked up to two hops upstream of central complex input neurons (Extended Data Fig. 2). We excluded neurons intrinsic to the central complex. For upstream partners, we included neurons that had at least five synapses with their downstream partner. After finding all neurons one to two hops upstream of the central complex input neurons, we determined which of them contained dendrites within optic lobe neuropils. This number of hops was selected to match the number of hops of the direct pathway from MeTu to TuBu to ER neurons. When calculating optic lobe weights, we summed the relative proportion of synapses that came from those optic lobe neurons. Renders of alternate pathways included the central complex neurons, upstream neurons that are either optic lobe neurons or neurons with upstream optic lobe partners, and those upstream optic lobe partners. The renders only contained these neurons if they accounted for â¥1% of the total non-central complex synaptic weight of their postsynaptic partner.
Mapping medulla columns to ommatidia coordinates
We used microCT data53 to assign each medulla column to an ommatidia. We determined the directions of ommatidia to the world as previously described53. Separately, we counted the numbers of R1âR6 photoreceptors in the lamina to identify the equatorial medulla columns, which have seven or eight photoreceptors compared to six in non-equatorial columns (data not shown). We then mapped ommatidia and medulla columns to separate hexagonal grids. Finally, using the equator as a reference, we aligned these hexagonal grids by minimizing unmatched points, hence assigning each medulla column a viewing (or sampling) direction in the visual field.
ER neuron visual area
For each âvisual columnâ of a given ER neuron, we calculate the values for the direct pathway as the sum of all weighted branches connecting the ER neuron via TuBu and MeTu neurons to the âvisual columnâ (Fig. 4a, middle). Each branch is the product of synaptic weights of TuBu neuron to ER neuron connection, MeTu neuron to TuBu neuron connection and MeTu neuron medulla column occupancy. MeTu neuron medulla column occupancy is calculated as the fraction of presynaptic sites closed to the column. The values for the indirect pathway are the sum of all weighted branches connecting the ER neuron via TuBu, TuTuB and MeTu neurons to the âvisual columnâ (Fig. 4b, middle). We overlaid the values for the direct and the indirect pathways with different colours (Fig. 4c,e).
Hemibrain comparison
The hemibrain dataset contains the entire central complex of the D. melanogaster brain, but only extends to include the AOTU of the left hemisphere. Therefore, it contains two sets of ER neurons, one set of TuBu neurons and only the boutons of one set of MeTu neurons. The MeTu neurons were named MC64 or MC61.
We used the Python module neuprint-python to look at the MC61 and MC64 that are presynaptic to the previously defined TuBu neurons. We first distinguished MeTu1âMeTu4 on the basis of their respective TuBu types. All MC64 are MeTu4, but a small population of MeTu4 was MC61 instead. We plotted the number of synapses within the lobula among MC61 and MC64 to determine that this distinction was due to differences in the number of lobula connections (data not shown). We distinguished MeTu4 in the same way as for FAFB, in which fewer than 15 synapses in the lobula denoted MeTu4d. We classified all other MeTu subtypes using their downstream TuBu partners. The only classification we were unable to make was that of MeTu3a and MeTu3b, because they were separated using upstream connections in the medulla, which the dataset did not include. We labelled these neurons MeTu3ab, and adjusted the FAFB one in comparison plots. After classification, there are 127 MeTu1, 39 MeTu2a, 14 MeTu2b, 64 MeTu3ab, 86 MeTu3c, 68 MeTu4a, 13 MeTu4b, 41 MeTu4c and 17 MeTu4d.
After obtaining all AVP neurons in FlyWire and neuprint, we compared the relative numbers of neurons among four hemispheres with ER and bihemispheric neurons, and three hemispheres with TuBu and MeTu neurons. We noticed discrepancies among TuBu and ER neuron counts, so we compared the ratios of ER to TuBu counts in the three hemispheres (Extended Data Fig. 9e,f).
Histology
To support the EM-based cell typing and to provide genetic tools for future studies of MeTu neurons, we report several split-GAL4 lines with preferential expression in subsets of MeTu types. These split-GAL4 lines were generated before the EM work; that is, are the output of an independent, light-microscopy-based effort to characterize MeTu cell types but are newly reported here. Driver lines and their candidate EM matches are listed in Supplementary Table 2. Figures show processed images displayed using VVD viewer (https://github.com/JaneliaSciComp/VVDViewer/releases). Lines and original image data are available at https://splitgal4.janelia.org/cgi-bin/splitgal4.cgi. Some lines are not currently maintained as stocks but can be reconstructed from the AD and DBD hemidrivers.
The general strategy and methods to generate and characterize these lines were as described for other split-GAL4 lines33,63. In brief, we searched for GAL4 lines with expression in MeTu cells using images of GAL4 driver expression patterns67,68,69, then screened the expression patterns of hemidriver (AD, DBD) combinations selected to target candidate MeTu types on the basis of these searches, and constructed stable fly lines for combinations with patterns of interest. Further characterization of these lines included imaging of overall expression patterns and, in most cases, of MCFO-labelled individual cells. Sample preparation and imaging, performed by the FlyLight Project Team using protocols available at https://www.janelia.org/project-team/flylight/protocols, were as in previous work34,70. For an overview of the Janelia FlyLight split-GAL4 project, see a previous study64. Some additional MeTu driver lines that are not included here are also available47.
Fly preparation for calcium imaging
All experiments were performed with seven-to-nine-day-old female flies. Blinding was impossible, and thus was not performed, because the morphology of the neurons is easily recognizable during imaging for each type of neurons. We did not perform sample size calculation but collected data from a fixed predetermined number of flies. Before experiments, flies were prepared as previously described6. In brief, a fly was anaesthetized on ice and transferred to a cold plate. The proboscis of the fly was pressed into the head and fixed with wax. In addition, the front and middle pairs of legs were removed. The fly was glued to a pin and positioned in a pyramid-shaped holder. We tilted the fly head 30â45° to the left and glued it to the holder with UV-curable gel. Next, we removed the cuticle on the head, together with ocelli and trachea, to expose the central brain for optical access. Muscle 16 was severed using a dissection needle to reduce brain movement. To further stabilize the brain and minimize motion, we covered the exposed brain with around 3âµl of saline with 3% low-gelling point agarose (Sigma-Aldrich), which was adopted from an imaging procedure of Drosophila Kenyon cells71. The brain was bathed in saline, as described in previous studies5, with an adjusted calcium concentration at 2.7âmM. The fly was then transferred to the microscope for recording.
Projector-based visual stimulation set-up
Visual stimuli were rear-projected onto a Teflon screen (0.254 mm thick, McMaster-Carr, item 8569K18) placed at the anterior-left-ventral side of the fly with a 45° inclination. We used a customized projector (Texas Instruments) with filters (AVR) to display blue stimuli with a wavelength peak at 450ânm. The stimuli were displayed at a frame rate of 60âHz and a resolution of 1,028âÃâ960. A photodiode was placed on the edge of the screen to detect a small flashing square for synchronization between the visual stimuli and calcium activity72. Visual stimuli were drawn and displayed by Psychtoolbox-3.
Two-photon calcium imaging
We used a custom-built two-photon scanning microscope (Janelia MIMMS 2.0) with a 40à objective (Nikon, NA 0.8, 3.5âmm WD). We used a Chameleon laser tuned to 930ânm with a maximum power of 10âmW for excitation and detected fluorescence by a GaAsP photomultiplier tube. We imaged the superior bulb over 15 focal planes, each separated by 1âµm, acquired at a volume rate of about 10âHz at a resolution of 128âÃâ128.
Calcium image analysis
All data processing and analyses were performed in MATLAB. We corrected for brain movement in xy directions by registering individual frames to the reference image using a cross-correlation algorithm. We generated reference images in Fiji by summing up images across the time series. We manually identified individual microglomeruli as an ROI on the basis of the videos of fluorescence changes, and we subtracted the mean fluorescence of an empty ROI from the same frame for each frame to compensate for noise. We defined the baseline fluorescence (F0) as the lowest 10% signal throughout the experiment. Because each microglomerulus does not occupy the entire depth of 15 planes in a volume, we averaged fluorescence from three consecutive planes and used the strongest value among 13 values in each volume as F, to calculate the ÎF/F0.
Receptive field analysis
The fly was presented with a square-shaped dot for 1âs followed by 1âs of darkness. The dot was randomly chosen from a set of 38 pre-indexed dots, which were 18° by 18° each in size and were not spatially overlapping. Each dot was tested for ten trials. In each trial, we calculated the response by subtracting the mean ÎF/F during the 1-s stimulation period from the mean ÎF/F during the 500âms before a dot appeared. We used the average response over ten trials to plot the receptive field. If the average response had either a P value higher than 0.05 in the Wilcoxon signed-rank test or a change smaller than the response of an empty ROI, we considered it unresponsive. We quantified the properties of an excitatory receptive field by fitting an ellipse into it by using the regionprops function in MATLAB.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.