Construction of spatially invariant connectome from local reconstructions
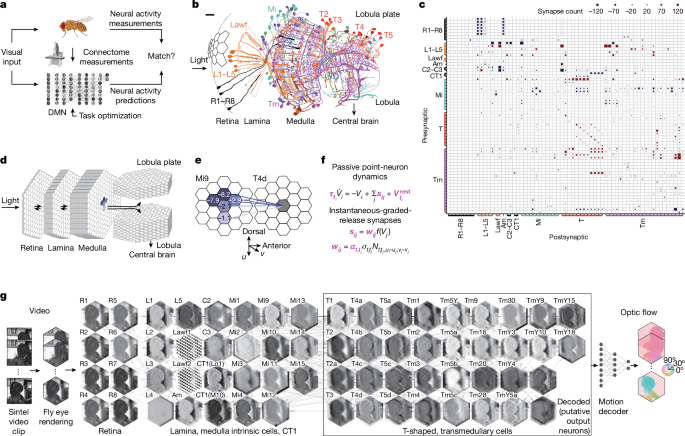
We built a computational model of the fly visual system that is consistent with available connectome data1,2,3,4,5, has biophysically plausible neural dynamics, and can be computationally trained to solve an ethologically relevant behavioural task, namely the estimation of optic flow. To achieve this, we developed algorithms to blend annotations from two separate datasets by transforming, sanitizing, combining and pruning the raw datasets into a coherent connectome spanning all neuropils of the optic lobe (Supplementary Note 1 and Supplementary Data files 1â3).
The original data stem from focused ion beam scanning electron microscopy datasets from the FlyEM project at Janelia Research Campus. The FIB-25 dataset volume comprises seven medulla columns and the FIB-19 dataset volume comprises the entire optic lobe and, in particular, detailed connectivity information for inputs to both the T4 and T5 pathways2,3,4. The data available to us consisted of 1,801 neurons, 702 neurons from FIB-25 and 1,099 neurons from FIB-19. For about 830 neurons, the visual column was known from hand annotation. These served as reference positions. Of the 830 reference positions, 722 belong to neuron types selected for simulation. None of the T5 cells, whose directional selectivity we aimed to elucidate, was annotated. We therefore built an automated, probabilistic expectation maximization algorithm that takes synaptic connection statistics, projected synapse centre-of-mass clusters and existing column annotations into account. We verified the quality of our reconstruction as described in Supplementary Note 1. Only the neurons consistently annotated with both 100% and 90% of reference positions used were counted to estimate the number of synapses between cell types and columns, to prune neuron offsets with low confidences.
Synaptic signs for most cell types were predicted on the basis of known expression of neurotransmitter markers (primarily the cell-type-specific transcriptomics data from ref. 30). For a minority of cell types included in the model, no experimental data on transmitter phenotypes were available. For these neurons, we used guesses of plausible transmitter phenotypes. To derive predicted synaptic signs from transmitter phenotypes, we assigned the output of histaminergic, GABAergic and glutamatergic neurons as hyperpolarizing and the output of cholinergic neurons as depolarizing. In a few cases, we further modified these predictions on the basis of distinct known patterns of neurotransmitter receptor expression (see ref.â30 for details). For example, output from R8 photoreceptor neurons, predicted to release both acetylcholine and histamine, was treated as hyperpolarizing or depolarizing, respectively, depending on whether a target cell type is known to express the histamine receptor gene ort (which encodes a histamine-gated chloride channel).
Representing the model as a hexagonal convolutional neural network
Our end-to-end differentiable61 DMN model of the fly visual system can be interpreted as a continuous-time neural ordinary differential equation62 with a deep convolutional recurrent neural network63 architecture that is trained to perform a computer vision task using backpropagation through time64,65. Our goal was to optimize a simulation of the fly visual system to perform a complex visual information processing task using optimization methods from deep learning. One hallmark of visual systems that has been widely exploited in such tasks is their convolutional nature66,67,68,69 (that is, the fact that the same computations are applied to each pixel of the visual input). To model the hexagonal arrangement of photoreceptors in the fly retina, we developed a hexagonal convolutional neural network (CNN) in the widely used deep learning framework PyTorch21 (ignoring neuronal superposition70), which we used for simulation and optimization of the model. We model columnar cell types, including retinal cells, lamina monopolar and wide-field cells, medulla intrinsic cells, transmedullary cells and T-shaped cells, as well as amacrine cells. The model comprises synapses from all neuropils and downstream- and upstream-projecting connections from the retina, lamina and medulla.
Neuronal dynamics
In detail, we simulated point neurons with voltages Vi of a postsynaptic neuron i, belonging to cell type ti using threshold-linear dynamics, mathematically equivalent to commonly used formulations of firing-rate models71
$${\tau }_{{t}_{i}}{\dot{V}}_{i}=-\,{V}_{i}+\sum _{j}{s}_{ij}+{V}_{{t}_{i}}^{\text{rest}}+{e}_{i}.$$
(1)
Neurons of the same cell type share time constants, \({\tau }_{{t}_{i}}\), and resting potentials, \({V}_{{t}_{i}}^{\text{rest}}\). Dynamic visual stimuli were delivered as external input currents ei to the photoreceptor (R1âR8), for all other cell types, eiâ=â0. In our model, instantaneous graded synaptic release from presynaptic neuron j to postsynaptic neuron i is described by
$${s}_{{ij}}={w}_{{ij}}\,f({V}_{j})={\alpha }_{{t}_{i}{t}_{j}}{\sigma }_{{t}_{i}{t}_{j}}{N}_{{t}_{i}{t}_{j}\varDelta u\varDelta v}\,f({V}_{j}),$$
(2)
comprising the anatomical filters in terms of the synapse count from electron microscopy reconstruction, \({N}_{{t}_{i}{t}_{j}\varDelta u\varDelta v}\), at the offset location \(\varDelta u={u}_{i}-{u}_{j}\) and \(\varDelta v={v}_{i}-{v}_{j}\) in the hexagonal lattice between two types of cells, \({t}_{i}\) and \({t}_{j}\), and further characterized by a sign, \({\sigma }_{{t}_{i}{t}_{j}}\in \{\,-\,1,+1\}\), and a non-negative scaling factor, \({\alpha }_{{t}_{i}{t}_{j}}\).
The synapse model entails a trainable non-negative scaling factor per filter that is initialized as
$${\alpha }_{{t}_{i},{t}_{j}}=\frac{0.01}{{\rm{\langle }}{N}_{{t}_{i},{t}_{j}}{{\rm{\rangle }}}_{\varDelta u,\varDelta v}},$$
with the denominator describing the average synapse count of the filter. Synapse counts, \({N}_{{t}_{i}{t}_{j}\varDelta u\varDelta v}\) from the connectome, and signs, \({\sigma }_{{t}_{i}{t}_{j}}\) from the neurotransmitter and receptor profiling, were kept fixed. The scaling factor was clamped during training to remain non-negative.
Moreover, at initialization, the resting potentials were sampled from a Gaussian distribution
$$\begin{array}{c}{V}_{{t}_{i}}^{\text{rest}} \sim {\mathcal{N}}\left({\mu }_{{V}^{\text{rest}}},{\sigma }_{{V}^{\text{rest}}}^{2}\right)\end{array}$$
with mean \({\mu }_{{V}^{\text{rest}}}=0.5\) (a.u.) and variance \({\sigma }_{{V}^{\text{rest}}}^{2}=0.05\) (a.u.). The time constants were initialized at \({\tau }_{{t}_{i}}=50\,{\rm{ms}}\). The 50 task-optimized DMNs were initialized with the same parameter values. During training, in Euler integration of the dynamics, we clamped the time constants as \({\tau }_{i}=\max \left({\tau }_{i},\varDelta t\right)\), so that they remain above the integration time step Ît at all times.
In total, the model comprises 45,669 neurons and 1,513,231 synapses, across two-dimensional (2D) hexagonal arrays 31 columns across. The number of free parameters is independent of the number of columns: 65 resting potentials, 65 membrane time constants, 604 scaling factors; and connectome-determined parameters: 604 signs and 2,355 synapse counts. Thus, the number of free parameters in the visual system model is 734.
In the absence of connectome measurements, the number of parameters to be estimated is much larger. With Tâ=â65 cell types (counting CT1 twice for the compartments in the medulla and lobula) and Câ=â721 cells per type for simplicity, the number of cells in our model would be TCâ=â46,865. Assuming a recurrent neural network with completely unconstrained connectivity and simple dynamics \({\tau }_{i}{V}_{i}=-{V}_{i}+{\sum }_{j}{w}_{{ij}}\,f({V}_{j})+{V}_{i}^{{\rm{rest}}}\), we would have to find \({({TC})}^{2}+2({TC})\,=\,\)\(2,196,421,955\) free parameters. Assuming a convolutional recurrent neural network with shared filters between cells of the same postsynaptic type, shared time constants and shared resting potentials, the amount of parameters reduces markedly to \({T}^{2}C+2T=3,046,355\). Further assuming the same convolutional recurrent neural network but additionally that convolutional filters are constrained to Fâ=â5 visual columns (that is, the number of presynaptic input columns in the hexagonal lattice is \(P=3F\left(F+1\right)+1\)), the amount of parameters reduces to \({T}^{2}P+2T=384,605\). Assuming as in our connectome that only Qâ=â604 connections between cell types exist, this reduces the number of parameters further to \({QP}+2T=55,185\). Instead of parametrizing each individual synapse strength, we assume that synapse strength is proportional to synapse count from the connectome times a scalar for each filter, reducing the number of parameters to \(Q+2T=734\) while providing enough capacity for the DMNs to yield realistic tuning to solve the task.
Convolutions using scatter and gather operations
For training the network, we compiled the convolutional architecture specified by the connectome and the sign constraints to a graph representation containing: a collection of parameter buffers shared across neurons and/or connections; a collection of corresponding index buffers indicating where the parameters relevant to a given neuron or connection can be found in the parameter buffers; and a list of pairs (presynaptic neuron index, postsynaptic neuron index) denoting connectivity. This allowed us to efficiently simulate the network dynamics through Euler integration using a small number of element-wise, scatter and gather operations at each time step. We found that this is more efficient than using a single convolution operation or performing a separate convolution for each cell type as each cell type has its own receptive fieldâsome much larger than othersâand the number of cells per type is relatively small.
Optic flow task
Model training
An optic flow field for a video sequence consists of a 2D vector field for each frame. The 2D vector at each pixel represents the magnitude and direction of the apparent local movement of the brightness pattern in an image.
We frame the training objective as a regression task
$$\hat{{\bf{Y}}}\left[n\right]={\rm{Decoder}}\left({\rm{DMN}}\left({\bf{X}}\left[0\right],…,{\bf{X}}\left[n\right]\right)\right),$$
with \(\hat{{\bf{Y}}}\) being the optic flow prediction, and X being the visual stimulus sequence from the Sintel dataset, both sampled to a regular hexagonal lattice of 721 columns. With the objective to minimize the square error loss between predicted optic flow and target optic flow fields, we jointly optimized the parameters of both the decoder and the visual system network model described above.
In detail, for training the network, we added randomly augmented, greyscaled video sequences from the Sintel dataset sampled to a regular hexagonal lattice of 721 columns to the voltage of the 8 photoreceptor cell types (Fig. 1f and equation (1)). We denote a sample from a minibatch of video sequences as \({\bf{X}}\in {{\mathbb{R}}}^{N,C}\), with N being the number of time steps, and C being the number of photoreceptor columns. The dynamic range of the input lies between 0 and 1. Input sequences during training entailed 19 consecutive frames drawn randomly from the dataset and resampled to match the integration rate. At the original frame rate of 24âHz, this corresponds to a simulation of 792âms. We did not find that an integration time step smaller than 20âms (that is, a frame rate of 50âHz after resampling) yielded qualitatively superior task performance or more realistic tuning predictions. We interpolated the target optic flow in time to 50âHz temporal resolution. To increase the amount of training data for better generalization, we augmented input and target sequences as described further below. At the start of each epoch, we computed an initial state of the networkâs voltages after 500âms of grey stimulus presentation to initialize the network at a steady state for each minibatch during that epoch. The network integration for a given input X results in simulated sequences of voltages \({\bf{V}}\in {{\mathbb{R}}}^{N,{T}_{C}}\), with TC being the total number of cells. The subset of voltages, \({{\bf{V}}}_{\text{out}}\in {{\mathbb{R}}}^{N,D,C}\), of the D cell types in the black rectangle in Fig. 1g was passed to a decoding network. For decoding, the voltage was rectified to avoid the network finding biologically implausible solutions by encoding in negative dynamic ranges. Furthermore, it was mapped to Cartesian coordinates to apply PyTorchâs standard spatial convolution layers for decoding and on each time step independently. In the decoding network, one layer implementing spatial convolution, batch normalization, softplus activation and dropout, followed by one layer of spatial convolution, transforms the D feature maps into the 2D representation of the estimated optic flow, \(\hat{{\bf{Y}}}\in {{\mathbb{R}}}^{N,2,C}\).
Using stochastic gradient descent with adaptive moment estimation (β1â=â0.9, β2â=â0.999, learning rate decreased from 5âÃâ10â5 to 5âÃâ10â6 in ten steps over iterations, batch size of four) and the automatic gradient calculation of the fully differentiable pipeline, we optimized the biophysical parameters through backpropagation through time such that they minimize the L2-norm between the predicted optic flow, \(\hat{{\bf{Y}}}\), and the ground-truth optic flow, Y:
$$L({\bf{Y}},\widehat{{\bf{Y}}})=\parallel {\bf{Y}}-\widehat{{\bf{Y}}}\parallel .$$
We additionally optimized the shared resting potentials for 150,000 iterations, using stochastic gradient descent without momentum, with respect to a regularization function of the time-averaged responses to naturalistic stimuli of the central column cell of each cell type, tcentral, to encourage configurations of resting potentials that lead to non-zero and non-exploding activity in all neurons in the network. We weighted these terms independently with γâ=â1, encouraging activity greater than a, and δâ=â0.01, encouraging activity less than a. We chose \({\lambda }_{V}=0.1\) and aâ=â5 in arbitrary units. With B being the batch size and T being the number of all cell types, the regularizer is
$$R(V)=\frac{{\lambda }_{V}}{BT}\sum _{b}\sum _{{t}_{{\rm{central}}}}\{\begin{array}{cc}\gamma {(\bar{V}-a)}^{2}, & {\rm{if}}\,\bar{V}=\frac{1}{N}\sum _{n}{V}_{b{t}_{{\rm{central}}}}[n]\le a\\ \delta {(\bar{V}-a)}^{2}, & {\rm{if}}\,\bar{V} > a.\end{array}$$
We regularly checkpointed the error measure \(L\left({\bf{Y}},\hat{{\bf{Y}}}\right)\) averaged across a held-out validation set of Sintel video clips. Models generalized on optic flow computation after about 250,000 iterations, yielding functional candidates for our fruit fly visual system models that we analysed with respect to their tuning properties.
Task-optimization dataset
We optimized the network on 23 sequences from the publicly available computer-animated film Sintel12. The sequences have 20â50 frames, at a frame rate of 24 frames per second and a pixel resolution of 1,024âÃâ436. The dataset provides optical flow in pixel space for each frame after the first of each sequence. As the integration time steps we use are faster than the actual sampling rate of the sequences, we resample input frames accordingly over time and interpolate the optic flow.
Fly-eye rendering
We first transformed the RGB pixel values of the visual stimulus to normalized greyscale between 0 and 1. We translated Cartesian frames into receptor activations by placing simulated photoreceptors in a 2D hexagonal array in pixel space, 31 columns across resulting in 721 columns in total, spaced 13 pixels apart. The transduced luminance at each photoreceptor is the greyscale mean value in the 13âÃâ13-pixel region surrounding it.
Augmentation
We used: random flips of input and target across one of the three principal axes of the hexagonal lattice; random rotation of input and target around its six-fold rotation axis; adding element-wise Gaussian noise with mean zero and variance \({\sigma }_{n}=0.08\) to the input X (then clamped at 0); random adjustments of contrasts, \(\log c\sim {\mathcal{N}}(0,{\sigma }_{c}^{2}=0.04)\), and brightness, \(b\sim {\mathcal{N}}\left(0,{\sigma }_{b}^{2}=0.01\right)\), of the input with \({X}^{{\prime} }=c\left(X-0.5\right)+0.5+{cb}\).
In addition, we âstridedâ the fly-eye rendering across the rectangular raw frames in width, subsampling multiple scenes from each. We ensured that such subsamples from the same scene were not distributed across training and validation sets. Input sequences in chunks of 19 consecutive frames were drawn randomly in time from the full sequences.
Black-box decoding network
The decoding network is feedforward, convolutional and has no temporal structure. Aspects of the architecture are explained in the section entitled Model training. The spatial convolutions have a filter size of 5âÃâ5. The first layer transforms the Dâ=â34 feature maps to an eight-channel intermediate representation, which is further translated by an additional convolutional layer to a three-channel intermediate representation of optic flow. The third channel is used as shared normalization of each coordinate of the remaining 2D flow prediction. The decoder uses PyTorch-native implementations for 2D convolutions, batch normalization, softplus activation and dropout. We initialized its filter weights homogeneously at 0.001.
Model characterization
Task error
To rank models on the basis of their task performance, we computed the standard optic flow metric of average end-to-end point error (EPE)72, which calculates the average over all time steps and pixels (that is, here columns) of the error
$${\rm{EPE}}({\bf{Y}},\hat{{\bf{Y}}})=\frac{1}{{NC}}\sum _{n}\sum _{c}\sqrt{{({y}_{1c}[n]-{\hat{y}}_{1c}[n])}^{2}+{({y}_{2c}[n]-{\hat{y}}_{2c}[n])}^{2}}$$
between predicted optic flow and ground-truth optic flow and averaged across the held-out validation set of Sintel sequences.
Importance of task optimization and connectome constraints
We generated DMNs with different constraints to assess their relative importance for predicting tuning properties. First, we studied the importance of task optimization of DMN parameters. We created 50 DMNs with random Gaussian-distributed parameters, and task-optimized only their decoding network, to obtain baseline values for both the task error and the accuracy of predicting tuning curves without task optimization of the DMN.
In the full DMN, we constrained single synapses by connectome cell-type connectivity, cell connectivity, synapse counts and synapse signs (equation (2)) and task-optimized the non-negative type-to-type unitary synapse scaling factor \({{\boldsymbol{\alpha }}}_{{t}_{i},{t}_{j}}\). Next, we trained five additional task-optimized DMNs with different connectome constraints (Fig. 2d and Extended Data Fig. 2aâd).
In these five additional types of DMN, we additionally task-optimized the terms in bold, rather than using connectome measurements, related to synaptic currents from a presynaptic cell j to a postsynaptic cell i: known single-cell connectivity, unknown synapse count: \({w}_{{ij}}={\sigma }_{{t}_{i},{t}_{j}}{{\bf{m}}}_{{t}_{i},{t}_{j},\varDelta u,\varDelta v}\), in which \({{\bf{m}}}_{{t}_{i},{t}_{j},\varDelta u,\varDelta v}\) is non-negative; known cell-type connectivity, unknown single-cell connectivity and synapse counts: \({w}_{i,j}={\sigma }_{{t}_{i},{t}_{j}}{{\bf{m}}}_{{t}_{i},{t}_{j},-3 < \varDelta u,\varDelta v,\varDelta u+\varDelta v < 3}\) (that is, for all connected cell types, a connection weight was learned for all cells up to a distance of three columns in hexagonal coordinates, with known signs); known single-cell connectivity and synapse counts, but unknown synapse signs: \({w}_{i,j}={{\boldsymbol{\alpha }}}_{{t}_{i},{t}_{j}}{{\boldsymbol{\sigma }}}_{{t}_{j}}{N}_{{t}_{i},{t}_{j},\varDelta u,\varDelta v}\) (that is, connection weights were fixed by measurements, but signs optimized); known single-cell connectivity, but unknown synapse signs and synapse counts: \({w}_{i,j}={{\bf{w}}}_{{t}_{i},{t}_{j},\varDelta u,\varDelta v}\), (that is, all non-zero connection weights were optimized, including their signs); or known cell-type connectivity, unknown single-cell connectivity, synapse counts and synapse signs: \({w}_{i,j}={{\bf{w}}}_{{t}_{i},{t}_{j},-3 < \varDelta u,\varDelta v,\varDelta u+\varDelta v < 3}\) (that is, for all connected cell types, a connection weight and sign was learned for all cells up to distance of three columns). We trained 50 models per DMN type. The task-optimized parameters in each case are highlighted using bold symbols. We randomly initialized the models with \({{\bf{m}}}_{{t}_{i},{t}_{j}},{{\bf{w}}}_{{t}_{i},{t}_{j}}\sim {\mathcal{N}}\left(0,\frac{2}{{n}_{{\rm{in}}}}\right)\), in which \({n}_{{\rm{in}}}\) is the number of cell connections and m is non-negative, and \({{\boldsymbol{\sigma }}}_{{t}_{j}}\in \{\,-\,1,1\}\) with equal probability.
Unconstrained CNN
We trained unconstrained, fully convolutional neural networks on the same dataset and task to provide an estimate of a lower bound for the task error of the DMN. Optic flow was predicted by the CNN from two consecutive frames
$$\begin{array}{c}\hat{Y}\left[n\right]={\rm{CNN}}\left(X\left[n\right],X\left[n-1\right]\right).\end{array}$$
with the original frame rate of the Sintel film. We chose 5 layers for the CNN with 32, 92, 136, 8 and 2 channels, respectively, and kernel size 5 for all but the first layer, for which the kernel size is 1. Each layer performs a convolution, batch normalization and exponential linear unit activation, except the last layer, which performs only a convolution. We optimized an ensemble of 5 unconstrained CNNs with 414,666 free parameters each using the same loss function, \(L\left(Y,\hat{Y}\right)\), as for the DMN. We used the same dataset (that is, hexagonal sequences and augmentations from Sintel) for training and validating the CNN as that used for training and validating the DMN, enabled by two custom modules mapping from the hexagonal lattice to a Cartesian map and back.
Circular flash stimuli
To evaluate the contrast selectivity of cell types in task-optimized models, we simulated responses of each DMN to circular flashes. The networks were initialized at an approximate steady state after 1âs of grey-screen stimulation. Afterwards the flashes were presented for 1âs. The flashes with a radius of 6 columns were ON (intensity Iâ=â1) or OFF (Iâ=â0) on a grey (Iâ=â0.5) background. We integrated the network dynamics with an integration time step of 5âms. We recorded the responses of the modelled cells in the central columns to compute the FRI.
FRI
To derive the contrast selectivity of a cell type, \({t}_{i}\), we computed the FRI as
$${{\rm{FRI}}}_{{{\rm{t}}}_{{\rm{i}}}}\begin{array}{c}=\frac{{r}_{{{\rm{t}}}_{\text{central}}}^{{\rm{peak}}}\left(I=1\right)-{r}_{{{\rm{t}}}_{\text{central}}}^{{\rm{peak}}}\left(I=0\right)}{{r}_{{{\rm{t}}}_{\text{central}}}^{{\rm{peak}}}\left(I=1\right)+{r}_{{{\rm{t}}}_{\text{central}}}^{{\rm{peak}}}\left(I=0\right)}\end{array}$$
from the non-negative activity
$$\begin{array}{c}{r}_{{{\rm{t}}}_{\text{central}}}^{{\rm{peak}}}\left(I\right)=\mathop{\max }\limits_{n}{V}_{{t}_{\text{central}}}\left[n\right]\left(I\right)+\left|\mathop{\min }\limits_{n,I}{V}_{{t}_{\text{central}}}\left[n\right]\left(I\right)\right|,\end{array}$$
from voltage responses \({V}_{{t}_{\text{central}}}\left[n\right]\left(I\right)\) to circular flash stimuli of intensities \(I\in \{0,1\}\) lasting for 1âs after 1âs of grey stimulus. We note that our index quantifies whether the cell depolarizes to ON- or to OFF-stimuli. However, cells such as R1âR8, L1 and L2 can be unrectified (that is, sensitive to both light increments and light decrements), which is not captured by our index.
For the P values reported in the results, we carried out a binomial test with probability of correct prediction 0.5 (H0) or greater (H1) to test whether both the median FRI from the DMN ensemble and the task-optimal model can predict the contrast preferences. Additionally, we found for each individual cell type across 50 DMNs that predictions for 29 out of 31 cell types are significant (Pâ<â0.05, binomial).
Moving-edge stimuli
To predict the motion sensitivity of each cell type in task-constrained DMNs, we simulated the response of each network, initialized at an approximate steady state after 1âs of grey-screen stimulation, to custom generated edges moving to 12 different directions, \(\theta \in [{0}^{^\circ },{30}^{^\circ },{60}^{^\circ },{90}^{^\circ },{120}^{^\circ },{150}^{^\circ },{180}^{^\circ },{210}^{^\circ },{240}^{^\circ },{270}^{^\circ },\)\({300}^{^\circ },{330}^{^\circ }]\). We integrated the network dynamics with an integration time step of 5âms. ON-edges (Iâ=â1) or OFF-edges (Iâ=â0) moved on a grey (Iâ=â0.5) background. Their movement ranged from â13.5° to 13.5° visual angle and we moved them at six different speeds, ranging from 13.92°âsâ1 to 145°âsâ1 (\(S\in [{13.92}^{^\circ }\,{{\rm{s}}}^{-1},{27.84}^{^\circ }\,{{\rm{s}}}^{-1},{56.26}^{^\circ }\,{{\rm{s}}}^{-1},{75.4}^{^\circ }\,{{\rm{s}}}^{-1},\)\({110.2}^{^\circ }\,{{\rm{s}}}^{-1},{145.0}^{^\circ }\,{{\rm{s}}}^{-1}]\)). In Fig. 2d, we report the correlation between predicted motion-tuning curves to the single experimentally measured tuning curve. We take the maximum correlation across six investigated speeds to make the correlation measure robust to potential variations in preferred speeds.
DSI
We computed a DSI73 of a particular type \({t}_{i}\) as
$$\begin{array}{r}{{\rm{DSI}}}_{{{\rm{t}}}_{{\rm{i}}}}(I)=\frac{1}{|{\mathbb{S}}|}\sum _{S\in {\mathbb{S}}}\frac{|{\sum }_{\theta \in \varTheta }{r}_{{{\rm{t}}}_{{\rm{central}}}}^{{\rm{peak}}}({\rm{I}},{\rm{S}},\theta )\exp (i\theta )|}{\mathop{\text{max}}\limits_{I\in {\mathbb{I}}}|{\sum }_{\theta }{r}_{{{\rm{t}}}_{{\rm{central}}}}^{{\rm{peak}}}({\rm{I}},{\rm{S}},\theta )|}\end{array}$$
from rectified peak voltages
$$\begin{array}{r}{r}_{{{\rm{t}}}_{{\rm{central}}}}^{{\rm{peak}}}({\rm{I}},{\rm{S}},\theta )=\mathop{\text{max}}\limits_{n}{V}_{{t}_{{\rm{central}}}}^{+}[n]({\rm{I}},{\rm{S}},\theta ),\end{array}$$
elicited from moving-edge stimuli. We rectify the voltage to quantify the tuning of the effective output of the cell, and to avoid the denominator becoming zero. We parameterized movement angle \(\theta \in \varTheta \), intensities \(I\in {\mathbb{I}}\), and speeds \(S\in {\mathbb{S}}\) of the moving edges. To take the response magnitudes into account for comparing the DSI for ON- and for OFF-edges, we normalized by the maximum over both intensities in the denominator. To take different speeds into account, we averaged over \({\mathbb{S}}\).
Normalization of model neural activity for averaging across models in a cluster
Threshold-linear networks have arbitrary units for the voltages and currents. Therefore, we normalized the neural activity before averaging the neural activity predictions from different models. For a single cell or cell type t, we first divided responses (voltages or rectified voltages) by the root mean square across the cellâs responses to the naturalistic stimuli:
$$\begin{array}{c}{r}_{t}^{{\rm{| }}{\rm{| }}\cdot {\rm{| }}{\rm{| }}\sim 1}[n]=\frac{{r}_{t}[n]}{| | \frac{1}{{SN}}{{\bf{R}}}_{t}^{{\rm{nat}}.}{{\rm{| }}{\rm{| }}}_{2}},\end{array}$$
in which \({{\bf{R}}}_{t}^{{\rm{nat}}.}\in {{\mathbb{R}}}^{S,N}\) is the cellâs response vector to S sequences from the Sintel dataset with N time steps and \({r}_{t}\left[n\right]\) is the cellâs response to any stimuli. This normalization makes averages (Fig. 4a,b,dâe and Extended Data Figs. 4, 7, 8, and 9a,b) independent to variation in the scale of neural activity from model to model. We normalized input currents equivalently (Fig. 4b and Extended Data Figs. 4, 7, and 8) by the same normalization factor. We exclude solutions for which the denominator becomes zero.
Determining whether a cell type with asymmetric inputs counts as direction selective
We counted a cell type as direction selective if the DSIs from its synthetic measurements were larger than 99% of DSIs from non-motion selective cell types (that is, those with symmetric filters). We note, however, that estimates of the spatial asymmetry of connectivity from existing connectome reconstructions can be noisy.
For deriving the 99% threshold, we first defined a distribution \(p\left({d}^{* }|{{\rm{t}}}_{{\rm{sym}}}\right)\) over the DSI for non-direction-selective cells, from peak responses to moving edges of cell types with symmetric inputs, tsym. We computed that distribution numerically by sampling
$$\begin{array}{r}{d}^{* }=\frac{|{\sum }_{{\theta }^{* }}{r}_{{{\rm{t}}}_{{\rm{central}}}}^{{\rm{peak}}}({\rm{I}},{\rm{S}},{\theta }^{* })\exp i\theta |}{|{\sum }_{\theta }{r}_{{{\rm{t}}}_{{\rm{central}}}}^{{\rm{peak}}}({\rm{I}},{\rm{S}},\theta )|}\end{array}$$
for 100 independent permutations of the angle \({\theta }^{* }\). We independently computed \({d}^{* }\) for all stimulus conditions, models and cell types with symmetric inputs. From \(p\left({d}^{* }|{{\rm{t}}}_{{\rm{sym}}}\right)\), we derived the threshold \({d}_{{\rm{thresh}}}=0.357\) as the 99% quantile of the random variable \({d}^{* }\), meaning that the probability that a realization of \({d}^{* } > {d}_{{\rm{thresh}}}\) is less than 1% for cell types with symmetric inputs. To determine whether an asymmetric cell type counts as direction selective, we tested whether synthetically measuring direction selectivity larger than dthresh in that cell type is binomial with probability 0.1 (H0) or greater (H1). We identified 12 cell types with asymmetric inputs (T4a, T4b, T4c, T4d, T5a, T5b, T5c, T5d, TmY3, TmY4, TmY5a and TmY18) as direction selective (Pâ<â0.05) from our models, and 7 cell types with asymmetric inputs to not count as direction selective (T2, T2a, T3, Tm3, Tm4, TmY14 and TmY15; see Extended Data Fig. 5 as reference for cell types with symmetric and asymmetric inputs).
Uniform manifold approximation and projection and clustering
We first simulated central column responses to naturalistic scenes (24âHz Sintel video clips from the full augmented dataset) with an integration time step of 10âms. We clustered models in feature space of concatenated central column responses and sample dimension. Next, we computed a nonlinear dimensionality reduction to two dimensions using the UMAP (uniform manifold approximation and projection) algorithm, and fitted Gaussian mixtures of 2 to 5 components, with the number of components that minimize the Bayesian information criterion, using the Python libraries umap-learn and scikit-learn38,74.
Single-ommatidium flashes
To derive spatio-temporal receptive fields of cells, we simulated the response of each network to single-ommatidium flashes. Flashes were ON (Iâ=â1) or OFF (Iâ=â0) on a grey (Iâ=â0.5) background and presented for [5, 20, 50, 100, 200, 300]âms after 2âs of grey-screen stimulation and followed by 5âs of grey-screen stimulation.
Spatio-temporal, spatial and temporal receptive fields
We derived the spatio-temporal receptive field (STRF) of a cell type \({t}_{i}\) as the baseline-subtracted responses of the central column cell to single-ommatidium flashes \(J\left(u,v\right)\) at ommatidium locations \(\left(u,v\right)\):
$${{\rm{STRF}}}_{{t}_{{\rm{central}}}}[n](u,v)={V}_{{t}_{{\rm{central}}}}[n](\,J(u,v))-{V}_{{t}_{{\rm{central}}}}[n=0](\,J(u,v)).$$
We derived spatial receptive fields (SRFs) from the responses to flashes (20âms in Fig. 4d) \(J\left(u,v\right)\) at the point in time at which the response to the central ommatidium impulse is at its extremum:
$${\rm{SRF}}\left(u,v\right)={\rm{STRF}}\left(n={{\rm{argmax}}}_{n}\left|{\rm{STRF}}\left[n\right]\left(0,0\right)\right|,u,v\right).$$
We derive temporal receptive fields (TRFs) from the response to a flash \(J\left(0,0\right)\) at the central ommatidium: \({\rm{TRF}}\left[n\right]={\rm{STRF}}\left[n\right]\left(0,0\right)\). For averaging receptive fields across multiple models, we first normalize the voltages as described above.
Maximally excitatory naturalistic and artificial stimuli
First, we found the naturalistic maximally excitatory stimulus, \({\text{X}}^{* }\), by identifying the Sintel video clip, X, from the full dataset with geometric augmentations that elicited the highest possible response in the central column cell of a particular cell type in our models.
$${{\rm{X}}}^{* }=\mathop{{\rm{argmax}}}\limits_{{\rm{X}}\in {\rm{Sintel}}}{V}_{{t}_{{\rm{central}}}}\left({\rm{X}}\right).$$
Next, we regularized the naturalistic maximally excitatory stimulus, to yield \({\text{X}}^{{\prime} }\), capturing only the stimulus information within the receptive field of the cell, with the objective to minimize
$$L({\rm{X}}{\prime} )=\sum _{n}\parallel {V}_{{t}_{{\rm{central}}}}({{\rm{X}}}^{* })[n]-{V}_{{t}_{{\rm{central}}}}({\rm{X}}{\prime} )[n]{\parallel }^{2}+\frac{1}{C}\sum _{c}\parallel {\rm{X}}{\prime} [n,c]-0.5{\parallel }^{2}.$$
The first summand preserves the central response to \({\text{X}}^{* }\), and the second regularizes the irrelevant portions of the stimulus outside the receptive field to grey (Iâ=â0.5).
In addition, we computed artificial maximally excitatory stimuli75.
Model selection
To describe the most data-consistent motion tuning mechanisms predicted by the ensemble at the level of single-cell currents, for Extended Data Figs. 4, 7 and 8, we automatically selected those models from the ensemble with tuning matching to empirical data. Specifically, we selected models with correct contrast tuning in the respective target cells and their inputs (Fig. 4c and Extended Data Fig. 3d), with the DSI larger than the threshold \({d}^{* }\) derived above, and with a correctly predicted preferred direction (45° acceptance angle, assuming 225° for TmY3).
Training synthetic connectomes
Training feedforward synthetic ground-truth connectome networks
Sparsified feedforward neural networks with six hidden layers (linear transformations sandwiched between rectifications) with equal number of neurons in each hidden layer functioned as ground-truth connectome networks. The main results describe networks with 128 neurons per hidden layer. We interpret the individual units as neurons with voltage
$${V}_{i}=\sum _{j}{s}_{ij}+{V}_{i}^{{\rm{rest}}}=\sum _{j}{\sigma }_{j}{c}_{ij}{m}_{ij}\,f({V}_{j})+{V}_{i}^{{\rm{rest}}},$$
with presynaptic inputs \({s}_{{ij}}\) and resting potentials \({V}_{i}^{\text{rest}}\). The connectome-constrained synapse strength, \({w}_{{ij}}\), is characterized by the adjacency matrix \({c}_{{ij}}\), the signs \({\sigma }_{j}\), and the non-negative weight magnitudes \({m}_{{ij}}\). \({c}_{{ij}}=1\) if the connection exists, else \({c}_{{ij}}=0\). To respect Daleâs law, the signs were tied to the presynaptic identity, j.
We identified the parameters \({\sigma }_{j}\), \({m}_{{ij}}\) and \({V}_{i}^{\text{rest}}\) by task optimization on handwritten digit classification (Modified National Institute of Standards and Technology (MNIST) database)76. We determined adjacency matrices, \({c}_{{ij}}\), for a given connectivity percentage using an iterative local pruning technique, the lottery ticket hypothesis algorithm77. The algorithm decreases the connectivity percentage of the ground-truth connectome networks while maintaining high task accuracy.
We optimized the ground-truth connectome networks and all simulated networks described below in PyTorch with stochastic gradient descent with adaptive moment estimation (ADAM with AMSGrad), learning rate 0.001, batch size 500, and an exponentially decaying learning rate decay factor of 0.5 per epoch. To constrain the weight magnitudes to stay non-negative, we clamped the values at zero after each optimization step (projected gradient descent). The parameters after convergence minimize the cross-entropy loss between the predicted and the ground-truth classes of the handwritten digits. More implementation detail is available in Supplementary Note 5.
Simulated networks with known connectivity and unknown strength
Simulated networks inherited connectivity, \({c}_{{ij}}\), and synapse signs, \({\sigma }_{j}\), from their respective ground-truth connectome networks. In simulated networks, signs and connectivity were held fixed. Weight magnitudes, \({m}_{{ij}}\), and resting potentials, \({V}_{i}^{\text{rest}}\), were initialized randomly and task-optimized. Just like ground-truth connectome networks, simulated networks were trained on the MNIST handwritten digit classification task until convergence.
Simulated networks with known connectivity and known strength
Alternatively, we imitate measurements of synaptic counts from the ground-truth weight magnitudes:
$${\widetilde{m}}_{{ij}}={m}_{{ij}}{{\epsilon }}_{{ij}}\,{\rm{with}}\,{{\epsilon }}_{{ij}}\sim {\mathcal{U}}(1-\sigma ,1+\sigma ),$$
with multiplicative noise to imitate spurious measurements. We used \(\sigma =0.5\) for the main results. Weight magnitudes were initialized at the measurement, \({\widetilde{m}}_{{ij}}\), and task-optimized on MNIST with the additional objective to minimize the squared distance between optimized and measured weight magnitudes, \({\widetilde{m}}_{{ij}}\) (L2 constraint, Gaussian weight magnitude prior centred around the simulated networkâs initialization). We weighted the L2 constraint ten times higher than the cross-entropy objective to keep weight magnitudes of the simulated networks close to the noisy connectome measurements. Resting potentials, \({V}_{i}^{\text{rest}}\), were again initialized randomly and task-optimized.
Measuring ground-truth-simulated network similarity
Ground-truth-simulated network similarity was measured by calculating the median Pearsonâs correlation of tuning responses (rectified voltages) of corresponding neurons in the ground-truth-simulated network pair. In each of the 6 hidden layers, Nâ=â100 randomly sampled neurons were used for comparison. Response tuning was measured over input stimuli from the MNIST test set (Nâ=â10,000 images). Results are medians over all hidden layers and over 25 ground-truth-simulation network pairs.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.