Resource availability
Requests for additional information and reagents should be addressed to the lead author (C.R.). All data generated in this paper can be shared on request.
Fly husbandry and strains
Flies were reared at 25â°C or 30â°C for RNAi experiments, with 40â50% humidity on a standard cornmeal-agar food in a 12-h lightâdark cycle. Canton-S (CS) strain flies were used as wild type. Flies were sorted under CO2 anaesthesia within 6âh of emergence and housed in same-sex groups of 20, except for the males that were to be tested in the behavioural experiments, which were kept in groups of 4 per vial. Virgin females for the behavioural experiments were collected using the hs-hid conditional virginator transgenic line. L3 larvae were heat shocked at 37â°C for 1.5âh. Additional strains used and their sources61,62,63,64,65,66,67,68,69 are outlined in Supplementary Table 3.
Trans-retinal food
Trans-retinal (R2500-100MG, CAS number: 116-31-4, Sigma-Aldrich) was stored at â20â°C as a 50âmM stock solution diluted in ethanol and wrapped in foil. To blend retinal homogeneously into the food, 60âμl of stock solution was directly pipetted into 6-ml vials of liquid cornmeal-yeast food except for the experiment in Fig. 3e in which OvAbg flies were not exposed to food supplemented with trans-retinal factor.
Behaviour
Threat setup
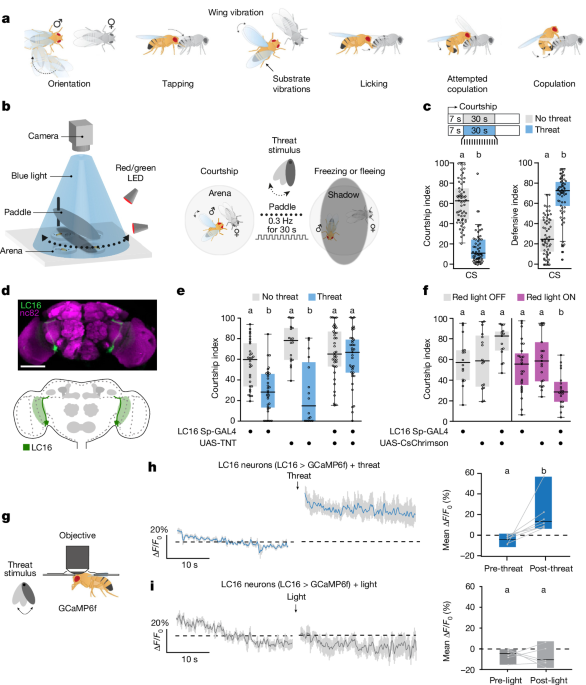
Experiments were recorded at 27 frames per second using a Mako U-130B camera mounted with an infrared filter (BP735-40.5, Midopt). The visual threat was generated by repeatedly passing a 13âcmâÃâ6âcmâÃâ2âcm 3D-printed opaque oblong paddle through a blue-light beam (455ânm). This created an overhead shadow at periodic intervals of 0.3âHz for 30âs. The paddle was set 5âcm above the courtship chambers (â 20âmm, 5âmm) at a 90o angle on a servo motor controlled by a custom-built Arduino code, which controlled the movement parameters of the paddle (frequency set to 0.3âHz and number of cycles set to 9). The mechanical threat was generated using a Sony XP500-X speaker playing a loud 3-Hz binaural beat (https://www.youtube.com/watch?v=Y-urmCRs61I&t=713s) causing surface vibrations. Courtship chambers were illuminated from the bottom using an infrared backlight.
Behavioural assays
Behavioural assays were conducted at 25â°C under continuous blue light between 09:00 and 13:00. Tested males were 5â7 days of age and transferred to fresh food vials 1 day before experiments. For males used in optogenetic assays, flies were transferred to food enriched with trans-retinal 3 days before the experiment. Vials containing retinal were wrapped in foil. Virgin females were decapitated and used within a maximum of 3âh to preserve chemical signature and motor reflexes during the experiment.
Action selection assay
The action selection assay presented a naive male coupled with a decapitated hs-hid virgin female with a choice between continuing to court the female or interrupting the ritual in response to the threat. The threat was delivered after consistent courtship of at least 7âs (early), 2âmin (middle) or 4âmin (late). Only males that started to court during the first 5âmin of the trial and until threat delivery were considered in the analysis. All assays were manually analysed using the behavioural analysis software BORIS70, and the following parameters were quantified to measure the effect of the threat on male courtship behaviour.
The courtship index is defined by the percentage of time (in seconds) the male spends courting the female over the total time of the threat delivery (30âs). We considered that males initiated courtship by demonstrating full wing extension and a persistent courtship behaviour of at least 7âs towards the female. We considered courtship as the display of stereotyped courtship events that include tapping of the female with the forelegs of the male, singing (wing extension and vibration), licking (male proboscis extension) and attempts at copulation in which the male bends the abdomen towards the female and attempts to mount her. See Fig. 1a for a schematical representation of these behaviours.
The defensive index is defined by the percentage of time (in seconds) the male spends displaying defensive behaviours (that is, escaping and freezing) over the total time of the threat delivery (30âs).
As a control, the behaviour was assessed in the absence of the visual threat during the same time window according to the same criteria.
Optogenetic assay
Flies were tested in a transparent circular chamber (â 20âmm, Hâ=â5âmm for courtship; and â 24âmm, Hâ=â3âmm for the locomotion assay) and illuminated from underneath with either 660-nm (red) or 515-nm (green) light in the absence or presence of the threat. Refer to Supplementary Table 4 for the optogenetic experimental conditions corresponding to each figure. The light was turned ON 1âs before the first threat passed.
Locomotion assay
Individual flies were introduced into a circular chamber (â 24âmm, Hâ=â3âmm) and left to acclimatize for 3âmin. After the acclimatation period, flies were subjected to the threat (9 cycles and frequency of 0.3âHz). The walking speed of the flies (thresholded at values larger than 4âmmâsâ1 to be considered as âwalkingâ) was assessed using the Ethovision XT17 software. The change in walking speed was calculated by subtracting the average walking speed of the 30âs after threat from the 30-s average before threat delivery.
Two-photon functional imaging
Tethered male flies (3â6 days of age) had their head capsules dissected in a sugar-free HL3-like saline-filled imaging chamber with a central hole (for details on fly dissection, see ref. 71). Flies were then placed under a multiphoton microscope (Femto2D-Resonant by Femtonics), and expressed either the calcium indicator GCaMP or GRABDA in different sets of neurons (see Supplementary Table 3 for details on genotypes). Fluorescence was generated by a Ti-Sapphire laser centred on 920ânm (Chameleon Ultra II, Coherent). Images with a pixel size of 0.3âÃâ0.3âμm were acquired with a Ã20, 1.0 NA water-immersion objective, controlled by the MESc v3.5 software (Femtonics). Fast recordings were taken at a speed of 30âHz with a resonant scan head using MESc software (Femtonics). Analysis was performed using NOSA software v1.1.16 (neuro-optical signal analysis)72 and a customized R script or Graphpad Prism, Regions of interest (ROIs) were manually drawn for analysis. Data were converted into tiff files and processed using a SavitzkyâGolay filter or moving average of 2âs when brain movement was strong (Figs. 4c and 5b). No baseline/photobleaching correction was applied to any of the imaging data. The final time resolution was 6âfps (Femtonics microscope data) or 2âfps (Optogenetic data from Nikon microscope). Mean intensity values were calculated as ÎF/F0 (in %), whereas F0 was defined as the mean F from baseline activity (first 30âs in Figs. 1h,i, 2i,j,m,n, 4e and 5c,d,h,k,l and Extended Data Figs. 3j, 4h, 7b,e; the first 20âs in Figs. 4c,f, and 5b and Extended Data Figs. 6fâi,k,l and 7a; the first 15âs in Fig. 5e,f and Extended Data Fig. 7e,g; and the first 2âs in Fig. 2e and Extended Data Fig. 3b).
Threat delivery under the two-photon microscope
The threat was delivered as previously described (see the âThreat setupâ section). The paddle and light source were placed below the microscope and inclined towards the chamber in a way that the passing shadow reached the tethered flyâs eye. Calcium signals in LC16 axons and PMPD neurons were recorded for 30âs before and 60âs immediately after the threat exposure (calculation windows in Figs. 1h,i and 5c,d,h,k,l: last 10âs before and first 10âs after; Fig. 4f and Extended Data Fig. 7e,g: last 15âs before and 30âs after). As LC16 neurons respond to laser onset, the first 2âs of each recording were excluded from the analysis. Conditions under the microscope were set to more than 20â°C and 40% humidity.
Application of serotonin or dopamine
100 µl of serotonin (H9523, Sigma-Aldrich) or dopamine (H8502, Sigma-Aldrich) diluted in sugar-free HL3 solution was applied directly onto the Drosophila brain through the open head capsule. The final concentration was 100âµM for serotonin and 500âµM for dopamine. Calcium signals were recorded 50âs before and 100 s immediately after application (first 30âs of pre-application and last 30âs of post-application were taken for quantification).
Courtship progression under the microscope
For examining courtship progression, 5â8-day-old virgin male flies were used. Flies were tethered and dissected as previously described, leaving legs and proboscis freely moveable (or fixed depending on the experiment indicated for each figure). Note that the fixation position of the male onto the imaging chamber does not allow for wing extension. Agitated males that did not stop moving for 10âs during the first 5âmin under the microscope were discarded. Immediately upon recording initiation, a decapitated 3â5-day-old virgin female tethered onto a moveable arm controlled by a micromanipulator was presented to the male with her abdomen oriented towards the head of the male fly. Following male contact with the female, calcium or GRABDA signals were recorded for a total duration of 4âmin, while the fly behaviour was simultaneously observed using a video camera (Thorlabs C1285R12M and SM1D12D iris diaphragm) recording at 7âfps. The first 20âs and last 20âs were taken for quantification (except Extended Data Fig. 6g: 1â20âs, 240â260âs and 400â420âs). Abdomen bending was manually analysed frame by frame. As tethered flies show typical behaviour that includes moving the abdomen back and forth, only full-bending events (the tip of the abdomen bending underneath the thorax) that lasted longer than 1âs or 6 frames were considered as part of courtship behaviour.
Optogenetic experiments during in vivo calcium imaging
Experiments were conducted using a Nikon A1R+ multiphoton microscope with a galvo scanner at a speed of 2âHz. We used the two-photon 1,040-nm red laser of the microscope to activate CsChrimson while simultaneously recording the calcium activity within the ROI (see the details for the conditions in the main text figure legends and Supplementary Table 4). To activate OvAbg neurons, experiments were carried out using a Femtonics microscope with the same imaging parameters mentioned previously. A 590-nm LED positioned below and towards the tethered fly was used for optogenetic activation of CsChrimson (15 or 7 repetitions of 1-s LED-on and 1-s LED-off intervals) while recording simultaneously. To activate PPM1/2 neurons during threat delivery, 15 repetitions of red light were used overlapping the 30âs of threat exposure under the microscope. LED stimulation artefacts were removed for clarity. As the acquisition was carried out continuously, the post-sequence shown in the graph displays the fluorescence intensity immediately after the LED stopped (Fig. 4d).
Focal dopamine injection
Fly preparation and imaging were conducted as described previously40 using a Nikon A1R+ multiphoton microscope. The sugar-free HL3-like saline was added with 30 units of Papain (Roche) and applied to the head capsule for 10âmin to digest the glial sheath of the brain and facilitate removal. Flies were subjected to local dopamine (10âmM diluted in saline) or saline injection via a micropipette (saline used for injection contained no CaCl2 or MgCl2). The injection solution was labelled with Texas Red (Invitrogen by Thermo Fisher Scientific, dextran, 10,000âMW) to visualize the pipette and the localization of the injections. Multiple (2â5) injections were given per experiment and averaged, resulting in a single average trace per experiment. Fluorescence traces were extracted using FIJI (ImageJ). F0 for the ÎF/F calculations was the average baseline fluorescence of the 10 frames immediately preceding the injection. Calculation windows for mean ÎF/F0 % was 10 s pre and last 10 s post. ROIs were selected manually.
Immunohistochemistry
Three-to-five-day-old male fly brains were dissected in ice-cold PBS and fixed in 4% paraformaldehyde solution at room temperature for 20âmin. Fixed brains were then washed four times in PBST (0.5%) for 30âmin and blocked with normal goat serum (5%) for 30â60âmin. The brains were then incubated with primary antibodies (anti-GFP chicken, 1:1,000 or 1:2,000, 13970, Abcam; anti-dsRed rabbit, 1:250, 632496, Takara; and nC82 anti-Brp, 1:50, DSHB) for 2â3 days at 4â°C. After four 20-min washes in PBST, the brains were incubated overnight with secondary antibodies (Alexa Fluor 488 goat anti-chicken IgG, 1:1,000 (A28175) or 1:2,000 (A32931), Thermo Fisher Scientific); Alexa Fluor 546 goat anti-mouse, 1:2000, A11018, Thermo Fisher; and Alexa Fluor 546 goat anti-rabbit, 1:2,000, A11071, Thermo Fisher). After four 20-min washes in PBST, brains were mounted in Vectashield on a glass slide before scanning with a Leica SP8 confocal microscope, a Nikon A1 confocal microscope or a Zeiss LSM900 with AiryScan2 module.
Split-GFP immunohistochemistry
Three-to-seven-day-old male fly brains were dissected in room temperature PBS and fixed in 4% paraformaldehyde solution at room temperature for 20âmin. Fixed brains were then washed in PBST (0.3%) three times for 20âmin each and blocked with normal goat serum (5%) for 30âmin. The brains were then incubated with anti-Brp (nC82, 1:50, DSHB) with 5% goat serum for 2 days at 4â°C. No anti-GFP antibody was used. After three 20-min washes in PBST, the brains were incubated with Alexa Fluor 546 goat anti-mouse (1:2,000, A11018, Thermo Fisher) for 2 days at 4â°C. After four 20-min washes in PBST, brains were mounted in Vectashield on a glass slide before scanning with a Nikon A1 confocal microscope.
Reconstituted split-GFP signal was quantified using ImageJ. The GFP signal was taken as the average pixel intensity within manually drawn volumes (freehand ROIs in multiple z-slices) around the LC16 axon terminals and cell bodies. The background fluorescence (from an ROI in a proximal brain region outside the LC16 neuron) was subtracted from the GFP signal. Statistical significance was evaluated by t-tests and two-way ANOVA in GraphPad Prism 9.
Connectomics search
We used the neuprint (hemibrain v1.2.1 dataset)39 platform to search for candidate neurons and subsequent connectivity (https://neuprint.janelia.org/).
-
Predicted link between LC16 and pC1a: Query Selection > General > Shortest paths > neuron Aâ=âLC16 # 1256830582 > Neuron Bâ=âpC1a # 359744514, Minimum weight = 3.
-
3D visualization of 5-HTPMPD01 and pC1 neurons: âdatasetâ:âhemibrain:v1.2.1â,âbodiesâ[â297230760â,â\n297908801â,â\n359744514â,â\n5813046951â,â\n267214250â,â\n267214250â,â\n392821837â,â\n359744514â,â\n5813046951â,â\n514850616â].
-
3D visualization of LC16 neurons and PPM1/2 neurons: âdatasetâ:âhemibrain:v1.2.1â,âbodiesâ[â1350945956â,â1288897930â,â1319927345â,â1319587380â,â1319579391â,â1254037524â,â1288893503â,â1289238972â,â1319586861â,â1319919918â,â1412989088â,â950229431â,â792040520â,â5813054384â].
Statistics and reproducibility
See Supplementary Tables 1 and 2 for details on statistics. All statistical tests were performed using R v2023.03.1â+â446 or GraphPad Prism 9. Each behavioural experiment was repeated at least three times over a minimum of 3 days. Individuals were tested only once. The sample size for the behavioural experiments always represents biologically independent animals. Behavioural indexes and calcium imaging quantification are displayed as boxplots. Boxes represent the lower (25th) and upper (75th) interquartile, respectively, and the horizontal line represents the median. Each dot on the plot represents a single fly. Courtship progression behavioural data and locomotion data do not follow a normal distribution, thus non-parametric MannâWhitney or KruskalâWallis tests, followed by a ConoverâIman multiple pairwise comparisons post-hoc test, have been applied on raw data (Pâ=â0.05, with a Bonferroni correction) for one factor experiments. To test the interaction between the genetic manipulations and the treatments, we applied two-way ANOVA. Significant differences are indicated by different letters at the level of Pâ<â0.05. We used a one-sample Wilcoxon signed-rank test (μâ=â0) to assess whether the speed change (â) in Extended Data Fig. 5e significantly deviated from 0. We indicated significance using an asterisk at the level of Pâ<â0.05.
Calcium imaging traces over time are represented as the mean âF/F0 (%; solid lines) with s.e.m. (shaded area). Quantification plots are shown as minimum/maximum plots and the median as the horizontal line. After verification of normality, a paired t-test or paired Wilcoxon signed-rank test was applied on mean âF/F0 (%) data from individual flies on specific time windows indicated in the figures and/or in the Methods. Significant differences are indicated by different letters (Pâ<â0.05). For inter-group comparisons, mean pre values were subsracted from mean post values and differences between genotypes and treatments were tested using one-way ANOVA, Kruskal-Wallis, t-test or Mann-Withney test as approriate. Experimenters were not blinded to the conditions of the experiments during data collection. Genotypes used for one experiment were tested simultaneously and in random order as well as random times during the day to avoid any influence of circadian timepoints and order of the experimental trials. We repeated all statistical tests excluding data points that were identified as outliers using the ROUT method in Prism with Qâ=â0.5%, and always obtained the same results, so we did not exclude outlier data points. Expression pattern of TH-C1-GAL4 and split-GAL4 lines, including LC16, P1, TRHR23E12 and plP10, were all imaged in nâ=â4 flies and were reliable across samples.
Randomization and blinding
Animals were never pre-assigned to a treatment or control group before the experiments. Behavioural and imaging experiments were performed in conjunction with their respective control cohorts. Randomization of animals was not implemented in this design.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.