Human participants
Both the patients and the relatives studied are alive and being followed up in Colombia. They were recruited by the Inborn Errors of Immunity group (formerly Primary Immunodeficiencies Group) in MedellÃn (Colombia). Written informed consent was obtained from the patients and relatives studied. This study was approved by the institutional ethics committees of the University of Antioquia (21-07-842), the Rockefeller University (JCA-0699) and INSERM (C10-07) and was performed in accordance with the local requirements of these institutions. Experiments on samples from human participants were conducted in the USA, France, Qatar and Colombia, in accordance with local regulations and with the approval of the institutional review boards of the corresponding institutions. Experiments on human lung tissue samples were approved by the regional institutional review board (Comité de Protection des Personnes Ãle de France VIII, Boulogne-Billancourt, France). Plasma samples from unrelated healthy individuals were collected at Sidra Medicine in accordance with a study protocol approved by the Clinical Research Ethics Board of Sidra Medicine, Qatar. Antibody profiles from selected, age-matched blood donors of diverse nationalities and individuals representative of the Arab general population were used for comparison (NCBI Sequence Read Archive: PRJNA685111 and PRJNA688708)60. Healthy volunteers for other studies were recruited in Colombia, France and the United States.
Extended case reports for P1 and P2
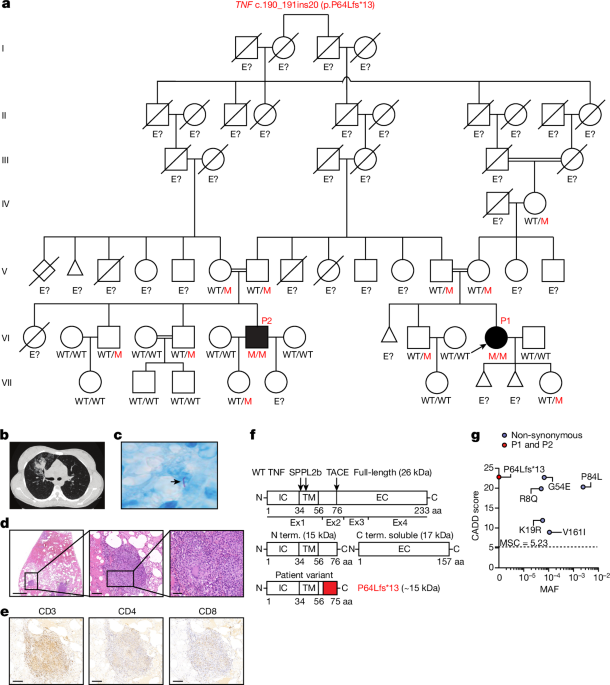
P1 was born in 1994. She was vaccinated with BCG vaccine strain Pasteur at birth with no documented adverse reaction. She received other vaccines in accordance with the national immunization program of Colombia, with no adverse effects. At the age of 19 years, she had a cough for a duration of one month, with fever (40â°C), pleuritic pain and unexplained weight loss. Chest X rays and CT scans revealed multiple pulmonary micronodules and a nodular lesion in the right lung. A segmental lobectomy of the right upper lobe was performed and revealed the presence of well-constituted, paucibacillary granulomas, some of which were necrotic. Immunohistochemistry detected the presence of CD3, CD4 and CD8 T cells, and epithelioid cells with a few giant cells. Bacterial cultures of sputum, bronchoalveolar lavage fluid and lung tissue were positive for a strain of M. tuberculosis that was found to be rifampicin-susceptible on Xpert PCR analysis. QuantiFERON and TST results were positive. Complete remission was achieved after 12 months of oral treatment with isoniazid, rifampicin, pyrazinamide and ethambutol. At 22 years of age, P1 was hospitalized at 28 weeks of pregnancy for a vaginal and urinary tract infection. She went into septic shock shortly afterwards, and an emergency caesarean section was required to extract the fetus, due to clinical chorioamnionitis. A blood culture was positive for L. monocytogenes, leading to the initiation of 7âdays of antimicrobial treatment with trimethoprim-sulfamethoxazole and ampicillin. The patient was discharged 10âdays after admission. Then, 5âmonths later, P1 was again diagnosed with pulmonary TB. Xpert PCR on bronchoalveolar lavage fluid again confirmed infection with rifampicin-susceptible M. tuberculosis. At the age of 22âyears, P1 was diagnosed with thymic hyperplasia, leading to thoracoscopy and complete thymectomy. Acid-fast bacillus (AFB) smear microscopy, Xpert PCR and cultures were negative for M. tuberculosis. A histological analysis of thymus tissue revealed mature adipose tissue, abundant thymic tissue with irregular islets but preserved architecture without signs of thymic carcinoma or thymoma. At the age of 23 years, P1 experienced a spontaneous abortion of unknown cause at 14 weeks of pregnancy. At the age of 27 years, she gave birth to a daughter. P1 is currently doing well without prophylactic treatment.
P2 was born in 1987 and is a first cousin of P1. He received all of the recommended vaccines according to the national vaccination schedule, including the live BCG vaccine (Pasteur strain), with no adverse effects. At the age of 18 years, he presented left pulmonary TB in the left lung with pleural effusion. P2 received 6âmonths of HRZE antimycobacterial treatment and responded well. However, he had a relapse eight months later and had to be hospitalized again. P2 was again diagnosed with TB, with positive QuantiFERON and TST results. He responded well to antimycobacterial treatment and is currently doing well off prophylactic treatment. Neither of the patients experienced any other severe infections caused by other bacteria, parasites or viruses. None of their relatives had a history of severe infectious diseases, including TB.
Cell lines
HEK293T cells (ATCC, CRL-11268, verified by the manufacturer via STR profiling) were cultured in Dulbeccoâs modified Eagle medium (DMEM) (Thermo Fisher Scientific, 11885084) supplemented with 10% fetal bovine serum (FBS). B cells from P1 and healthy controls were immortalized by infection with EBV and cultured in RPMI 1640 (Thermo Fisher Scientific, 11875093) supplemented with 10% FBS. HVS-T cells for P1 and healthy controls were generated with H. saimiri strain C488 for transformation or with the TERT transformation system. HVS-T cells were cultured in Panserin/RPMI 1640 (ratio 1:1) supplemented with 20% FBS, l-glutamine, gentamycin and 20âIUâmlâ1 human rIL-2 (Roche, 11147528001). Human iPS cells were maintained on mouse embryonic fibroblasts (MEFs) (Thermo Fisher Scientific, A34181) in knockout DMEM (Thermo Fisher Scientific, 10829-018) supplemented with 20% knockout serum replacement (Thermo Fisher Scientific, 10828-028), 2âmM l-glutamine (Thermo Fisher Scientific, 25030-024), 1% non-essential amino acids (Thermo Fisher Scientific, 11140-035), 1% penicillinâstreptomycin (Thermo Fisher Scientific; 15140-122), 0.2% β-mercaptoethanol (Thermo Fisher Scientific, 31350-010) and 10ângâmlâ1 basic fibroblast growth factor (bFGF, Peprotech, 100-18B). The cell lines were regularly tested and were found to be free of mycoplasma contamination.
Genetics
Genomic DNA from P1, P2 and their relatives was used for WES. Exome capture was performed using the SureSelect Human All Exon V4+UTR and SureSelect Human All Exon V6 kits (Agilent Technologies). Paired-end sequencing was performed on the HiSeq 2000 sequencer (Illumina) generating 100âbase reads. We used the Genome Analysis Software Kit (GATK) (v.3.4-46) best-practice pipeline to analyse our WES data. Reads were aligned with the human reference genome GRCh38 using BWA. PCR duplicates were removed using Picard tools v.3.1.1 (https://picard.sourceforge.net/). The GATK base quality-score recalibrator was used to correct sequencing artifacts. The GATK HaplotypeCaller v.4.1.4.1 was used to identify variant calls. Variants were annotated using SnpEff v.4.5 (https://snpeff.sourceforge.net/). Homozygosity rates were estimated from the patientsâ genomic DNA, as previously described61. Parametric multipoint linkage analysis was performed on the WES data using MERLIN v.1.1.2, assuming AR inheritance with complete penetrance and a damaging allele frequency of 1âÃâ10â4. Allele frequencies were estimated for 72,817 SNPs with the gnomAD v.2.1.1 American population. Markers were clustered with an r2 threshold (–rsq parameter) of 0.4. The genetic variant of interest was confirmed by PCR amplification of the region surrounding TNF exons 2 and 3 from the gDNA (5â²-AGCTGTTGAATGCCTGGAAGG-3â², and 5â²-CTCAGCGAGTCCTTCTCACATTG-3â²) followed by Sanger sequencing.
Detection of copy-number variants
We searched for copy-number variants in patient samples by applying ExomeDepth62 and HMZDelFinder_opt63 to WES samples mapped onto the human reference genome GRCh38. For both analyses, we selected the 50 nearest neighbours based on coverage as controls. We discarded HMZDelFinder_opt low-confidence candidates with a z score of above â1.2. All of the results were checked manually by read-mapping with Alamut Visual Plus v.1.5. The results are provided in Extended Data Table 3.
Phage immunoprecipitation sequencing
The reactivity of circulating antibodies against common pathogens in plasma samples from P1 and P2 and healthy controls was analysed by phage immunoprecipitationâsequencing (PhIPâseq) as previously described64. Pooled human plasma for IVIg (Privigen CSL Behring), human IgG-depleted serum (HPLASERGFA5ML, Molecular Innovations) and plasma samples from unrelated healthy adults were included as controls. PhIPâseq was performed with a modified version of the original VirScan phage library and data were processed as previously described60,65.
Detection of autoantibodies
Recombinant E. coli-derived IFNγ (285-IF-100/CF, R&D Systems), IL-12 (10018-IL-020, R&D Systems), IL-23 (1290-IL-010, R&D Systems) and TNF (300-01A, Peprotech) were first biotinylated with EZ-Link Sulfo-NHS-LC-Biotin (A39257, Thermo Fisher Scientific) according to the manufacturerâs instructions with a biotin-to-protein molar ratio of 1:12. The detection reagent contained a secondary antibody (Alexa Fluor 647 goat anti-human IgG (A21445, Thermo Fisher Scientific)) diluted in Rexxip F (P0004825, Gyros Protein Technologies; 1:500 dilution of the 2âmgâmlâ1 stock to yield a final concentration of 4âmgâmlâ1). PBS supplemented with 0.01% Tween-20 (0.01% PBS-T) and Gyros wash buffer (P0020087, Gyros Protein Technologies) were prepared according to the manufacturerâs instructions. Plasma samples were diluted 1:100 in 0.01% PBS-T and tested with the Bioaffy 1000 CD (P0004253, Gyros Protein Technologies) and a Gyrolab xPand (P0020520, Gyros Protein Technologies).
The blocking activity of anti-IFNα2, anti-IFNβ and anti-IFNÏ autoantibodies was determined using a reporter luciferase activity. In brief, HEK293T cells were transfected with a plasmid containing the Firefly luciferase gene under the control of the human ISRE promoter in the pGL4.45 backbone, and a plasmid constitutively expressing Renilla luciferase for normalization (pRL-SV40). Cells in DMEM supplemented with 2% FBS and 10% healthy control or patient serum/plasma (after inactivation at 56â°C, for 20âmin) were either left unstimulated or were stimulated with 100ângâmlâ1 IFNα2, IFNβ or IFNÏ at a concentration of 10ângâmlâ1 or 100âpgâmlâ1 for 16âh at 37â°C. Cells were lysed for 20âmin at room temperature and luciferase levels were measured using the Dual-Luciferase Reporter 1000 assay system (E1980 Promega).
Site-directed mutagenesis, transient and stable transfection
The pCMV3-SP-N-Myc-TNF vector was purchased from SinoBiological (HG10602-NM) and modified to remove the Myc tag sequence. Mutagenesis was performed with appropriate primers using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, 200521). HEK293T cells were transiently transfected in the presence of Lipofectamine 2000 (Thermo Fisher Scientific, 11668019). For stable transfection, WT and mutant TNF sequences were inserted into the pTRIP-CMV-Puro-2A vector (Addgene, 102611). For lentivirus production, HEK293T cells were transfected with 0.2âµg pCMV-VSV-G (Addgene, 8454), 0.2âµg pHXB2 (NIH-AIDS Reagent Program; 1069), 1âµg psPAX2 (Addgene plasmid 12260) and 1.6âµg pTRIP-CMV-Puro-2A in the presence of Lipofectamine 2000. After transfection for 6âh, the medium was replaced with 3âml DMEM supplemented with 10% FBS. The viral supernatant was collected 60âh later and concentrated with a Lenti-X Concentrator (Takara Bio, 631232). Concentrated lentivirus preparations were used for transduction in the presence of 10âµgâmlâ1 protamine sulfate (Merck, P3369).
Western blotting
Cell lines and primary cells were lysed with modified radioimmunoprecipitation assay (RIPA) buffer (25âmM TrisâHCl pHâ7.4, 150âmM NaCl, 1% NP-40 and 1âmM EDTA) supplemented with cOmplete ULTRA protease inhibitor cocktail tablets (Merck, 5892970001) and PhosSTOP tablets for phosphatase inhibition (Merck, 4906837001), 0.1âmM dithiothreitol (Thermo Fisher Scientific, 20290) and 1âmM PMSF (Merck, 10837091001). Protein samples were subjected to electrophoresis in 10â20% Criterion Tris-HCl Protein Gels (Bio-Rad, 3450043) or 4â20% TGX precast gels (Bio-Rad, 4561094), and the resulting bands were transferred onto Immobilon-P PVDF membranes (Millipore, IPVH00010). Membranes were probed with antibodies directed against TNF (C terminus, Abcam ab1793; and N terminus, Aviva Systems Biology, ARP80342_P050, both 1:1,000), GAPDH (Santa Cruz, sc-47724 and sc-25778 both 1:3,000), gp91phox (Santa Cruz, sc-130543, 1:300), p67phox (Merck, 07-002, 1:2,000), p47phox (Merck, 07-001, 1:5,000), p40phox (Merck, 07-503, 1:1,000), p22phox (Santa Cruz Biotechnology, sc-130550, 1:2,000) and EROS (Atlas Antibodies, HPA045696, 1:1,250) followed by incubation with secondary HRP-coupled anti-mouse (GE Healthcare, NXA931, 1:5,000) or anti-rabbit (GE Healthcare, NA934V, 1:5,000) antibodies. Images were analysed using Image Lab v.5.1 (Bio-Rad). Uncropped western blots are provided in Supplementary Fig. 1.
Deep immunophenotyping of leukocytes using spectral flow cytometry
Blood leukocyte subsets were phenotyped as previously described28. In brief, freshly thawed PBMCs were stained with LIVE/DEAD Fixable Blue (Thermo Fisher Scientific, L23105, 1:800 in PBS) and blocked by incubation with FcR Blocking Reagent (Miltenyi Biotec, 1:25) on ice for 15âmin. The cells were washed and surface stained with the following reagents on ice for 30âmin: Brilliant Stain Buffer Plus (BD Biosciences, 566385, 1:5), anti-γδTCR-BUV661 (BD Biosciences, 750019, 11F2, 1:50), anti-CXCR3-BV750 (BD Biosciences, 746895, 1C6, 1:20) and anti-CCR4-BUV615 (BD Biosciences, 613000, 1G1, 1:20) antibodies. Cells were then washed and surface stained by incubation with the following reagents on ice for 30âmin: anti-CD141-BB515 (BD Biosciences, 565084, 1A4, 1:40), anti-CD57-FITC (BD Biosciences, 347393, HNK-1, 3:250), anti-Vδ2-PerCP (BioLegend, 331410, B6, 3:500), anti-Vα7.2-PerCP-Cy5.5 (BioLegend, 351710, 3C10, 1:40), anti-Vδ1-PerCP-Vio700 (Miltenyi Biotec, 130-120-441, REA173, 1:100), anti-CD14-Spark Blue 550 (BioLegend, 367148, 63D3, 1:40), anti-CD1c-Alexa Fluor 647 (BioLegend, 331510, L161, 1:50), anti-CD38-APC-Fire 810 (BioLegend, 356644, HB-7, 3:100), anti-CD27-APC H7 (BD Biosciences, 560222, M-T271, 1:50), anti-CD127-APC-R700 (BD Biosciences, 565185, HIL-7R-M21, 1:50), anti-CD19 Spark NIR 685 (BioLegend, 302270, HIB19, 3:250), anti-CD45RA-BUV395 (BD Biosciences, 740315, 5H9, 3:250), anti-CD16-BUV496 (BD Biosciences, 612944, 3G8, 3:500), anti-CD11b-BUV563 (BD Biosciences, 741357, ICRF44, 1:100), anti-CD56-BUV737 (BD Biosciences, 612767, NCAM16.2, 3:250), anti-CD4-cFluor YG568 (Cytek, R7-20042, SK3, 3:250), anti-CD8-BUV805 (BD Biosciences, 612889, SK1, 3:250), MR1 tetramer-BV421 (NIH Tetramer Core Facility, 1:100), anti-CD11c-BV480 (BD Biosciences, 566135, B-ly6, 1:40), anti-CD45-BV510 (BD Biosciences, 563204, HI30, 3:250), anti-CD33-BV570 (BioLegend, 303417, WM53, 3:250), anti-iNKT-BV605 (BD Biosciences, 743999, 6B11, 1:25), anti-CD161-BV650 (BD Biosciences, 563864, DX12, 1:25), anti-CCR6-BV711 (BioLegend, 353436, G034E3, 3:250), anti-CCR7-BV785 (BioLegend, 353230, G043H7, 1:40), anti-CD3-Pacific Blue (BioLegend, 344824, SK7, 3:250), anti-CD20-Pacific Orange (Thermo Fisher Scientific, MHCD2030, HI47, 1:50), anti-CD123-Super Bright 436 (Thermo Fisher Scientific, 62-1239-42, 6H6, 1:40), anti-Vβ11-PE (Miltenyi Biotec, 130-123-561, REA559, 3:500), anti-CD24-PE-Alexa Fluor 610 (Thermo Fisher Scientific, MHCD2422, SN3, 1:25), anti-CD25-PE-Alexa Fluor 700 (Thermo Fisher Scientific, MHCD2524, 3G10, 1:25), anti-CRTH2-biotin (Thermo Fisher Scientific, 13-2949-82, BM16, 1:50), anti-CD209-PE-Cy7 (BioLegend, 330114, 9E9A8, 1:25), anti-CD117-PE-Dazzle 594 (BioLegend, 313226, 104D2, 3:250), anti-HLA-DR-PE-Fire 810 (BioLegend, L243, 1:50) and anti-CD66b-APC (eBioscience, 1305118, G10F5 1:50) antibodies. The cells were then washed and incubated with streptavidin-PE-Cy5 (BioLegend, 405205, 1:3,000) on ice for 30âmin. The cells were washed again, fixed by incubation in 1% PFA/PBS, washed again and acquired on the Aurora cytometer using SpectroFlo v.3.0 (Cytek). Subsets were manually gated using FlowJo v.10, and the results were visualized using R v.4.
Whole-blood stimulation for cytokine secretion
Whole-blood samples were collected into heparin-containing collection tubes. Blood samples were diluted 1:2 in RPMI 1640 and incubated with IFNγ (Imukin, Boehringer Ingelheim), IL-12 (20ângâmlâ1, R&D Systems, 219-IL), IL-23 (100ângâmlâ1, R&D Systems, 1290-IL), live Mycobacterium bovis BCG Pasteur substrain at a multiplicity of infection (MOI) of 20 or PMA (40ângâmlâ1, Sigma-Aldrich, P8139) and ionomycin (10âμM, Sigma-Aldrich, I9657). The supernatants were collected after 48âh and analysed using the LEGENDplex Human Inflammation Panel 1 (BioLegend, 740809).
PBMC stimulation assay with BCG and IL-23
For PBMC stimulation, 2âÃâ105 PBMCs were plated in 96-well, round-bottomed plates containing RPMI 1640 supplemented with 10% human serum and stimulated for 48âh with 2.5ângâmlâ1 IL-1β (R&D Systems, 201-LB), 50ângâmlâ1 IL-12 (R&D Systems, 219-IL), 100ângâmlâ1 IL-23 (R&D Systems, 1290-IL) and 0.1âv/v% BCG. PMA and ionomycin were added for the last 24âh. The supernatants were subjected to LEGENDplex multiplex ELISA with Human Inflammation Panel 1.
PBMC BCG stimulation assay
Freshly thawed PBMCs were dispensed into a 96-well round-bottomed plate at a density of 3âÃâ105 cells per well, in 200âμl per well lymphocyte medium. Cells were stimulated with live BCG at an MOI of 1, IL-12 (5ângâmlâ1, R&D Systems, 219-IL) or IL-23 (100ângâmlâ1, R&D Systems, 1290-IL). After 40âh of stimulation, GolgiPlug (BD Biosciences, 555029, 1:1,000) was added. The supernatants were collected after 8âh and evaluated with LEGENDplex Human Inflammation Panel 1. Cells were stained by incubation with Zombie NIR dye (BioLegend, 1:2,000) at room temperature for 15âmin, and were then incubated on ice for 30âmin with FcR blocking reagent (Miltenyi Biotec, 130-059-901, 1:50), 5-OP-RU-loaded MR1 tetramer-BV421 (NIH Tetramer Core Facility, 1:200), anti-CD3-V450 (BD Biosciences, 560365, UCHT1, 1:450), anti-CD4-BUV563 (BD Biosciences, 612912, SK3, 1:450), anti-CD8-BUV737 (BD Biosciences, 612754, SK1, 1:450), anti-CD20-BV785 (BioLegend, 302356, 2H7, 1:150), anti-CD56-BV605 (BioLegend, 362538, 5.1H11, 1:50), anti-γδTCR-Alexa Fluor 647 (BioLegend, 331214, B1, 1:50), anti-Vδ1-FITC (Miltenyi Biotec, 130-118-362, REA173, 1:150), anti-Vδ2-APC-Fire 750 (BioLegend, 331420, B6, 1:1350), anti-Vα7.2-Alexa Fluor 700 (BioLegend, 351728, 3C10, 1:50), anti-iNKT-BV480 (BD Biosciences, 746788, 6B11, 1:50) and anti-Vβ11-APC (Miltenyi Biotec, 130-125-529, REA559, 1:150) antibodies. Cells were fixed by incubation with 2% PFA/PBS on ice for 15âmin, and were then permeabilized/stained by incubation overnight at â20â°C in the perm buffer from the True-Nuclear Transcription Factor Buffer Set (BioLegend, 424401) with an intracellular cytokine panel: FcR blocking reagent, anti-IFNγ-BV711 (BioLegend, 502540, 4S.B3, 1:450), anti-TNF-BV510 (BioLegend, 502950, MAb11, 1:150), anti-IL-17A-PerCP-Cy5.5 (BioLegend, 512314, BL168, 1:1350), anti-T-bet-PE-Cy7 (BioLegend, 644823, 4B10, 1:1350) and anti-RORγT-PE (BD Biosciences, 563081, Q21-559, 1:50) antibodies. Cells were acquired on the Aurora cytometer (Cytek). Data were manually gated using FlowJo as previously described10,27 and then imported into R for further analysis. Cellular composition was visualized with uniform manifold approximation and projection based on the expression levels of CD3, CD4, CD8, CD20, CD56, γδTCR, Vδ1, Vδ2, Vα7.2, MR1, T-bet and RORγT, with the data downsampled to 10,000 cells per sample.
scRNA-seq analysis of leukocytes
Cryopreserved PBMCs from P1 and P2, one Colombian adult control individual and two patients with CYBB deficiency (one with MSMD and one with CGD) were analysed using scRNA-seq as previously described24. Thawed cells were washed with medium and filtered with a 70-µm-mesh MACS SmartStrainer (Miltenyi Biotec, 130-098-462). Cells were then washed three times with PBS plus 0.5% FBS and were finally filtered again with a 40âµm cell strainer (Corning, 352340) before capture on the 10x Genomics Chromium chip. Libraries were prepared with the Chromium Single Cell 3â Reagent Kit (v3 Chemistry) and sequenced on the Illumina NovaSeq 6000 sequencer (S1 flowcell). Sequences were preprocessed with CellRanger v.6 on the 10x Genomics Cloud Analysis platform (https://www.10xgenomics.com/products/cloud-analysis). Approximately 11,000 to 16,000 cells were captured per sample, with a mean of at least 24,000 reads per cell.
The data generated in this study were analysed together with historical controls from the laboratory, two previously reported adult controls and two patients with IRF1 deficiency7, and publicly available control PBMC datasets downloaded from the 10x Genomics web portal (https://support.10xgenomics.com/single-cell-gene-expression/datasets). Manually curated datasets were integrated with Harmony (v.3.8)66. Two runs of sequential graph-based clustering were performed. The second-round clustering focused on memory and effector T and NK cells to achieve cellular subset separation at the highest possible resolution. Clusters were manually identified with the aid of the SingleR v.2.60 pipeline67 guided by MonacoImmuneData68. The TotalSeq datasets from 10x were also used to determine the identity of each cluster. Principal component analysis was conducted on count data normalized through variance-stabilizing transformation (VST). Pseudobulk differential expression analysis was conducted using DESeq2 (v.1.40.2)69, excluding all public datasets. GSEA was conducted with the fgsea package v.1.30.0 by projecting the fold-change ranking onto the following MSigDB genesets (http://www.gsea-msigdb.org/gsea/msigdb/): H (Hallmark), C2 CP (Curated canonical pathways), C3 (Regulatory targets) and C5 (Gene ontologies). Intercellular communication analysis was performed using CellChat (v.1.5)70. All analyses were performed in R v.4 (http://www.R-project.org/)71.
Differentiation of MDMs
CD14+ cells were isolated from PBMCs with CD14 MicroBeads (Miltenyi Biotec, 130-050-201). GM-CSF-matured MDMs were generated with M1-Macrophage Generation Medium XF (PromoCell, C-28055). For the generation of MDMs in the presence of M-CSFâ+âIL-4, CD14+ cells were incubated for 7âdays in RPMIâ+â10% FCS supplemented with M-CSF (R&D Systems, 216-MC), and were then allowed to differentiate in the presence of M-CSFâ+âIL-4 (R&D Systems, 204-IL). The medium was replaced every 2 to 3 days.
Isolation of human lung macrophages
Lung tissue samples were obtained from patients undergoing surgical resection for suspected lung carcinoma without previous chemotherapy (nâ=â5), chronic aspergillosis in a context of bronchiectasis (nâ=â1) or explantation in the context of chronic respiratory insufficiency in a context of pulmonary amyloidosis (nâ=â1). A lung sample taken from a healthy area and considered to be surgical waste was dissected free of the pleura, visible airways and blood vessels72.
Lung macrophages were isolated using the adhesion method73. Fluid collected from the washing of minced peripheral lung tissues was centrifuged (2,000ârpm for 10âmin). The cell pellet was resuspended in RPMI medium with 10% FCS, 2âmM l-glutamine and antibiotics. The resuspended viable cells were then dispensed into 96-well plates. The plates were incubated for at least 1.5âh at 37â°C and non-adherent cells were removed by gentle washing. The adherent cells (approximately 3âÃâ104 cells per well) were >95% pure macrophages, as determined by MayâGrünwaldâGiemsa staining and CD68 immunocytochemistry. The day after isolation, macrophages were washed twice, and 100âµl RPMI medium supplemented with 1% FBS was added to each well.
Differentiation of AML cells
Monocytes were isolated from patients or healthy, unrelated controls and were differentiated into AML cells as previously described2,13,74. Monocytes were isolated from PBMCs using a classical monocyte isolation kit (Miltenyi Biotec, 130-117-337) and cultured in RPMI 1640 containing 10% human serum (H4522, Merck), 80âµgâmlâ1 poractant alfa (Curosurf, Chiesi) or 100âµgâmlâ1 Infasurf (ONY Biotec), 10ângâmlâ1 GM-CSF (BioLegend, 572914), 5ângâmlâ1 TGF-β (BioLegend, 781804) and 5ângâmlâ1 IL-10 (BD, 554611) for 6âdays. The surfactant and cytokines were replenished every other day.
ROS production assays
ROS production by neutrophils and monocytes was analysed by subjecting whole blood to red-blood-cell lysis and then incubating the lysate with dihydrorhodamine 123 (Thermo Fisher Scientific, D23806) in the presence or absence of PMA (400ângâmlâ1). Monocytes were labelled with a CD14 Pacific-Blue-conjugated antibody (BD, M5E2, 558121, 1:50). The analysis was performed on the LSRFortessa Cell Analyzer (BD). Superoxide production by MDMs, AMLs, lung macrophages and EBV-B cells was evaluated using the Superoxide Anion Assay Kit (Sigma-Aldrich, CS1000) according to the manufacturerâs instructions. In brief, 3âÃâ104 MDMs, AML cells or lung macrophages, or 1âÃâ105 EBV-B cells per well were plated in a 96-well plate in the presence or absence of superoxide dismutase and stimulated with PMA (400ângâmlâ1) or serum-opsonized heat-killed M. tuberculosis (InvivoGen, tlrl-hkmt-1). Luminescence was recorded on the Victor X4 plate reader. H2O2 production by MDMs, AML cells, lung macrophages and EBV-B cells was assessed using Amplex Red hydrogen peroxide (Thermo Fisher Scientific, A22188) in KrebsâRinger phosphate buffer. In brief, 3âÃâ104 cells per well were plated in 96-well flat-bottomed plates, stimulated for 24âh with 100âIUâmlâ1 IFNγ or 20ângâmlâ1 TNF (R&D Systems, 210-TA). H2O2 release was measured after stimulation with PMA or serum-opsonized heat-killed M. tuberculosis on the Victor X4 (Perkin Elmer) plate reader.
Immunofluorescence microscopy
Superoxide production by mitochondria was analysed using confocal microscopy with the MitoSOX Red Mitochondrial Superoxide Indicator (Thermo Fisher Scientific, M36008). We plated 4âÃâ105 cells GM-CSF-matured MDMs and AML cells from healthy controls on coverslips (80826, iBidi) and treated them with infliximab (5âµgâmlâ1) or isotype control (5âµgâmlâ1) for 48âh. Cells were stimulated with PMA (400ângâmlâ1), 0.5% DMSO or heat-killed M. tuberculosis (330âµgâmlâ1) for 4âh and were then incubated with 800ânM MitoSOX and 150ânM MitoTracker Green FM (Thermo Fisher Scientific, M7514) in HBSS buffer for 30âmin at 37â°C. Cells were washed with HBSS and stained by incubation for 5âmin with 1âµgâmlâ1 Hoechst 33342 (Thermo Fisher Scientific, 62249). Specific fluorescence was acquired with a Leica TCS SP8 STED (Leica) confocal laser-scanning microscope (Ã63 oil immersion lens).
Generation of KO iPS cell lines
iPS cell KO lines were generated by the Stem Cell Research facility at MSKCC. CRISPR sgRNAs targeting the gene of interest were designed using the algorithm available online (https://www.benchling.com/crispr/). The target sequences were inserted into the pX330-U6-Chimeric_BB-CBh-hSpCas9 vector (Addgene, 42230) for the generation of specific gene-targeting constructs. Variants were introduced into the parental C12 iPS cell line by electroporating single-iPS-cell suspensions (1âÃâ106 cells per reaction) with 4âµg sgRNA-construct plasmid in Nucleofector human stem cell solution (Lonza, VPH-5012). The electroporated cells were cultured for 4âdays in feeder-cell-free conditions and were then passaged at low density to obtain single-cell colonies. Individual colonies were picked 10âdays later, expanded and analysed using PCR. The lines obtained were karyotyped to check for the absence of chromosomal abnormalities.
Generation of iPS-cell-derived macrophages
We differentiated macrophages from human iPS cells as previously described75,76. In brief, iPS cells were maintained for three days in iPS cell medium (with 10ângâmlâ1 bFGF) and then 4âdays in iPS cell medium only. On day 7, iPS cell colonies were transferred to low-adhesion plates (Thermo Fisher Scientific, 657185) in the presence of 10âμM ROCK Inhibitor (Sigma-Aldrich, Y0503). After 6âdays of incubation, embryoid bodies were transferred to hematopoietic differentiation medium (APEL 2 medium (Stem Cell Tech, 05270) supplemented with 5% protein-free hybridoma (Thermo Fisher Scientific, 12040077), 1% penicillinâstreptomycin, 25ângâmlâ1 IL-3 (Peprotech, 200-03) and 50 ngâmlâ1 M-CSF (Peprotech, 300-25). From day 25 of differentiation onwards, cells in suspension were carefully collected, cultured for 6â10âdays in RPMI mediumâ+â10% FBS and 100ângâmlâ1 GM-CSF (Peprotech, 300-03).
L. monocytogenes infection
GM-CSF-matured macrophages were treated for 48âh with IFNγ (103âIUâmlâ1) and TNF (50ângâmlâ1). Infliximab or isotype control (both 5âµgâmlâ1) was added on days 1 and 7 of macrophage differentiation. Cells were infected with L. monocytogenes (MOI of 20 for 1âh at 37â°C). They were then washed twice and incubated with gentamicin (25âµgâmlâ1) for 30âmin, 3âh or 6âh. The cells were washed and lysed with 0.05% Triton X-100 in PBS and serial dilutions were plated on BHI agar and incubated overnight.
Infection of macrophages with M. tuberculosis
M. tuberculosis strain Erdman was used for the infection of AML cells. M. tuberculosis was grown to exponential growth phase in 7H9 Middlebrook medium supplemented with 0.5% glycerol, 0.05% Tween-80 and 10% oleic acid-albumin-dextrose-catalase (OADC), with shaking at 200ârpm. A single-cell mycobacterial suspension was prepared by washing bacteria collected by centrifugation twice with phosphate-buffered saline supplemented with 0.05% Tween-80 (PBST80) and then centrifuging at 200g for 10âmin. AML cells were infected at an MOI of 5, calculated from the optical density at 600ânm (OD600) of the final suspension, considering an OD600 of 1.0 to equate to approximately 5âÃâ108âCFU per ml. After 4âh, the cells were washed once with RPMI supplemented with 10% human serum and cultured for up to 5âdays.
Cytokine production and intracellular growth of M. tuberculosis in AML cells
The supernatant of M. tuberculosis-infected AML cells was passed through a filter with 0.22âµm pores twice and was then stored at â80â°C. Cytokine levels were quantified using LEGENDplex Human Inflammation Panel 1 (BioLegend, 740809) and LEGENDplex Human Cytokine Panel 2 (BioLegend, 741378). For the isolation of M. tuberculosis from infected AML cells, 0.01% Triton X-100 in sterile water was added to the wells and the plates were then incubated at room temperature for 20âmin. The culture was then passed up and down in a pipette to ensure AML cell lysis. Cell lysates were diluted with PBST80 and cultured on 7H10 Middlebrook agar supplemented with 0.5% glycerol and 10% OADC for 2âweeks at 37â°C. RNA was prepared by washing cell monolayers with PBS and then extracting the RNA in TRIzol.
RNA-seq analysis
We used the NovaSeq S1 platform (single-end, 100âbp reads), aiming to obtain 20â30âmillion reads per sample. The RNA-seq fastq raw data were inspected to ensure that they were of high quality and then mapped onto the human reference genome GRCh38 using STAR aligner (v.2.7)77. The aligned RNA-seq BAM files were used to quantify the gene-level read counts using featureCounts (v.1.6.0)78; these counts were then vst-normalized and log2-transformed using DESeq2 (v.1.40.2)69 to obtain values for the expression of all genes and all samples. The data were then analysed to identify the genes differentially expressed between the patients and the controls in baseline conditions without infection and at various timepoints (days 3 and 5) after stimulation (M. tuberculosis, M. tuberculosisâ+âTNF). We used the fgsea package v.1.30.0 to perform GSEA for MSigDB hallmark gene sets and gene ontologies.
Statistical analysis
All statistical analyses were performed using R v.4 (http://www.R-project.org/) and GraphPad Prism software v.9.5.0 (GraphPad). The statistical significance of quantitative differences between groups was assessed using MannâWhitney U-tests or unpaired two-tailed Studentâs t-tests. The statistical significance of differences between treated and untreated samples from the same donor was assessed using paired two-tailed Studentâs t-tests. P values are indicated only for statistically significant differences.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.