Participants
The study participants were 17 adult patients who were implanted with depth electrodes for seizure monitoring as part of an evaluation for treatment for drug-resistant epilepsy (Supplementary Table 1). No statistical methods were used to predetermine the sample size but this number of patients is large relative to other similar studies. All patients provided informed consent and volunteered to participate in this study. Research protocols were approved by the institutional review boards of Cedars-Sinai Medical Center, Toronto Western Hospital and the California Institute of Technology.
Psychophysical task and behaviour
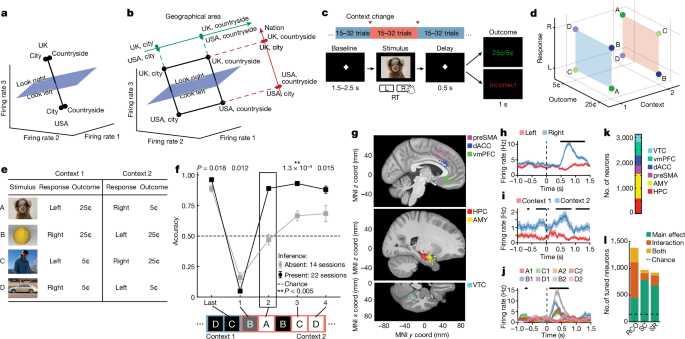
Participants performed a serial reversal learning task. There were two possible static stimulusâresponseâoutcome (SRO) maps, each of which was active in one of the two possible contexts. Context was latent and switches between context were uncued. Each recording session consisted of 280â320 trials grouped into 10â16 blocks of variable size (15â32 trials per block) with block transitions corresponding to a change in the latent context.
Patients completed 42 sessions of the task, typically in pairs of two back-to-back sessions on the same recording day (mean, 2.4 sessions per day, minimum two, maximum four; Supplementary Table 1). New stimuli were used in every session, thus requiring patients to re-learn the SRO maps through trial and error at the start of every session.
Each trial consisted of a blank baseline screen, stimulus presentation, speeded response from the participant, followed by feedback after a brief delay (Fig. 1a). Responses were either left or right in every trial. In each session, stimuli were four unique images, each chosen from a different semantic category (human, macaque, fruit, car). If a patient performed several sessions, new images not seen before by the patient were chosen for each session. The task was implemented in MATLAB (Mathworks) using PsychToolbox v.3.0 (ref. 58). Images were presented on a laptop positioned in front of the patient and subtended roughly 10° of visual arc (300âpx2, 1,024âÃâ768 screen resolution, 15.6âinch (40âcm) monitor, 50âcm viewing distance). Patients provided responses using a binary response box (RB-844, Cedrus).
Receipt of reward in a given trial was contingent on the accuracy of the response provided. In each trial, either a high or low reward (25 cents (¢) or 5¢) was given if the response was correct, and no reward (0¢) if incorrect. Whether a given trial resulted in high or low reward if the response was correct was determined by the fixed SRO map (Fig. 1c). Stimulusâresponse associations were constructed such that two out of four images (randomly selected) were assigned one response and the other two images were assigned the other (for example, human and fruit, left; macaque and car, right). Thus, in each context, each stimulus was uniquely specified by a combination of its correct response (left or right) and reward value (high or low). Crucially, the SRO maps of the two possible contexts were constructed so that they were the opposite of each other from the point of view of the associated response (Fig. 1c). To fully orthogonalize also associated reward, half of the reward values stayed the same and the others switched. This structured relationship of stimuli across contexts led to the full orthogonalization of the response, context and reward variables (Fig. 1b,c). Crucially, the stimulusâresponse map inversion across contexts provided the opportunity for patients to perform inferential reasoning about the current state of the SRO map, and therefore the latent context.
As rewards were provided deterministically, participants could infer that a context switch had occurred on receiving a single error to the first stimulus they encounter in the new context, and immediately respond correctly to the rest of the stimuli the first time they encounter them in the new context. The behavioural signature of inferential reasoning was thus the accuracy on the trials that occurred immediately after the first error trial. Specifically, we took a participantâs performance on the first instance of the other stimuli encountered in the new context as a measure of that participantâs inference capabilities (Extended Data Fig. 1a; note that although there are three inference trials after every context switch, each corresponding to a different stimulus, only the first inference trial was used to determine whether a session was in the inference present or absent group: below).
Patients completed several sessions of the task, in each of which new stimuli were chosen. After completion of the first session, the experimenter provided a standardized description of the latent contexts and SRO reversal to the patient (Supplementary Methods). These instructions were given regardless of how well the patient performed in the immediately preceding session. Each session took roughly 30âmin (mean 1,154âs, range 898â1,900âs), and the inter-session break during which instructions were provided lasted roughly 4âmin (Fig. 5a, bottom; mean duration 241âs, range 102â524âs). After this brief interlude, the participants completed the task again with a new set of four stimuli.
Behavioural control
We administered a control version of the task identical to the âfirst sessionâ described above to nâ=â49 participants recruited on Amazon Mechanical Turk (MTurk), who provided informed consent under a protocol approved by the institutional review board of Cedars-Sinai Medical Center (exempt study). No statistical methods were used to predetermine the sample size. We then used this data to calibrate the difficulty of the task. Most (roughly 75%) of the control participants demonstrated proper inference performance, and the remaining 25% demonstrating slow updating of SROs after a context switch, consistent with a behavioural strategy in which each stimulus is updated independently (Extended Data Fig. 1a).
Analysis of behaviour
Six of the 42 sessions were excluded due to at-chance performance in non-inference trials (binomial test, Pâ>â0.05). A session was classified as âinference presentâ if performance on the first of the three possible inference trials that occurred after the context switches was significantly above chance (timepoint 2 in Fig. 1f, binomial test on inference trial 1, Pâ<â0.05) and as âinference absentâ (nâ=â14 sessions, Pâ>â0.05, binomial test on inference trial 1) otherwise.
Electrophysiology
Extracellular electrophysiological recordings were conducted using microwires embedded within hybrid depth electrodes (AdTech Medical). The patients we recruited for this study had electrodes implanted in at least the hippocampus, as well as in addition subsets of amygdala, dorsal anterior cingulate cortex, upplementary motor area, ventromedial prefrontal cortex and VTC as determined by clinical needs (Supplementary Table 1). Implant locations were often bilateral, but some patients only had unilateral implants as indicated by clinical needs. Broadband potentials (0.1âHzâ9âkHz) were recorded continuously from every microwire at a sampling rate of 32âkHz (ATLAS system, Neuralynx). All patients included in the study had well-isolated single neuron(s) in at least one of the brain areas of interest.
Electrode localization was conducted using a combination of pre-operative magnetic resonance imaging and postoperative computed tomography using standard alignment procedures as previously described (using freesurfer v.5.3.0 and v.7.4.1)59,60. Electrode locations were coregistered to the to the MNI152-aligned CIT168 probabilistic atlas61 for standardized location reporting and visualization using Advanced Normalization Tools v.2.1 (refs. 59,60). Placement of electrodes in grey matter was confirmed through visual inspection of participant-specific computed tomography and magnetic resonance imaging alignment, and not through visualization on the atlas.
Spike detection and sorting
Raw electric potentials were filtered with a zero-phase lag filter with a 300âHzâ3âkHz passband. Spikes were detected and sorted using the OSort software package v.4.1 (ref. 62). All spike sorting outcomes were manually inspected and putative single units were isolated and used in all subsequent analyses. We evaluated the quality of isolated neurons quantitatively using our standard set of metrics63,64,65, including the proportion of inter-spike interval violations shorter than 3âms, signal-to-noise ratio of the waveform, projection distance between pairs of isolated clusters and isolation distance of each cluster relative to all other detected spikes. Only well-isolated neurons as assessed by these spike sorting quality metrics were included.
Selection of neurons, trials and analysis periods
Activity of neurons was considered during two epochs throughout each trial: the baseline period (base), defined as â1 to 0âs preceding stimulus onset on each trial and the stimulus period (stim), defined as 0.2 to 1.2âs following stimulus onset on each trial. Spikes were counted for every neuron on every trial during each of these two analysis periods. The resulting firing-rate vectors were used for all encoding and decoding analyses. For the stimulus period, because patients would sometimes respond before 1.2âs (reaction time 1.08â±â0.04âs over sessions), we determined that 75.15% of all spikes occurred before a response was provided across all recorded neurons, indicating that analyses performed with these spike counts predominantly, but not exclusively, reflect predecision processing. Tests of single-neuron selectivity were conducted using N-way ANOVAs with significance at Pâ<â0.05, where N was either 2 for models of stim ID (A, B, C, D) and context (1, 2), or 3 for models that included outcome (high, low), response (left, right) and context (1, 2). All variables were categorical, and all models were fit with all available interaction terms included. In Fig. 1l, a unit is marked as linearly tuned if it has at least one significant main effect, and nonlinearly tuned if it has at least one significant interaction term in the ANOVA model.
Population decoding analysis
Single-trial population decoding analysis was performed on pseudo-populations of neurons assembled across all neurons recorded across all patients. We pooled across sessions within each anatomically specified recording area as described previously59,66. We aggregated neurons across participants into a pseudo-population that consists of all neurons recorded in a given brain area, which allows us to examine populations of several hundred neurons in humans despite inability to record this many neurons simultaneously. This analysis approach is possible because all participants performed the same task, so that conditions could be matched across all relevant variables for a given trial in the pseudo-population (for example, trial 1 might be context 1, correct response, stimulus A, response right, outcome high). The justification for using this approach is threefold. First, independent population codes, in which the information that each neuron provides can be characterized by its own tuning curve, can be understood by recording one neuron at a time and aggregating them for analysis67. This is the type of code we are examining. Second, we seek to establish the content and structure of information that is reliably present in a given brain area across participants. This can only be achieved by recording in many participants. Third, in most instances, decoding from pseudo-populations yields the same results from simultaneously recorded neurons68,69. Results between the two approaches can differ when noise correlations are considered, which can have complex effects on the geometry of the underlying representation67. Here noise correlations are not the topic of interest. Noise correlations are present for the subgroups of neurons in the pseudo-population that were recorded simultaneously. To avoid potential effects of these remaining noise correlations, we removed them by randomly scrambling the order of trials for every neuron included in the pseudo-population (as we have described before59,66).
Decoding was conducted using support vector machines with a linear kernel and L2 regularization as implemented in MATLABâs fitcsvm function. No hyperparameter optimization was performed. All decoding accuracies are reported for decoding accuracy for individual trials. Decoding accuracy is estimated out of sample using fivefold cross-validation unless otherwise specified (for example, cross-condition generalization). Many of the decoding analyses in this work consist of grouping sets of distinct task conditions into classes, then training a support vector machine to discriminate between those two groups of conditions. Neurons included in the analysis were required to have at least K correct trials of every unique condition to be included in the analysis (Kâ=â15 trials unless otherwise stated). To construct the pseudo-population, we then randomly sampled K trials from every unique condition and divided those trials into the groups required for the current decoding analysis for every neuron independently. Randomly sampling correct trials in this way allowed us to destroy noise correlations that might create locally correlated subspaces from neurons recorded in the same area and session59.
To account for the variance in decoding performance that arose from this random subsampling procedure, all reported decoding accuracies are the average resulting from 1,000 iterations of subsampling and decoder evaluation. A similar trial balancing and subsampling procedure was conducted for all analyses that report decoding accuracy on incorrect trials, but with Kâ=â1 trial or condition required as incorrect for the neuron to be included in analysis. Various other analyses conducted throughout this work, including representation geometry measures, centroid distances and coding direction variances, all rely on this procedure of balanced correct and incorrect trial subsampling, and averaging across 1,000 iterations of the computed metric to study the relationships between task conditions in an unbiased manner. All reported values have been computed with this approach unless otherwise stated.
Balanced dichotomies
Our task has eight possible states (Fig. 1b). We characterized how neurons represented this task space by assessing how a decoder could differentiate between all possible âbalanced dichotomiesâ of these eight task conditions (Fig. 1b).
The set of all possible balanced dichotomies is defined by all possible ways by which the eight unique conditions can be split into two groups containing four of the conditions each (for example, four points in context 1 versus 4 points in context 2 is the context dichotomy). There are 35 possible balanced dichotomies (nchoosek(8,4)/2). We considered all 35 possible dichotomies to perform our analysis in an unbiased manner (Supplementary Table 2 shows all dichotomies). Some of the possible balanced dichotomies are easily interpretable because they correspond to variables that were manipulated in the task. We refer to these balanced dichotomies as the ânamed dichotomiesâ, which are: context, response, outcome, stimulus pair (stim pair) and parity. These dichotomies are shown individually in Extended Data Fig. 2. The stim pair dichotomy corresponds to the grouping of stimuli for which the response is the same in either context (A and C versus D and B; Fig. 2d). The parity dichotomy is the balanced dichotomy with the maximal nonlinear interaction between the task variables (Extended Data Fig. 2).
For decoding balanced dichotomies during the prestimulus baseline, the task states are defined by the values of the previous trial (not the upcoming, the identity of which is unknown to the participant). The reason for doing so is to examine persistent representations of the previous trial.
Defining decoding difficulty of dichotomies
We quantify the relative degree of nonlinear variable interactions needed by a neural population to classify a given dichotomy using a difficulty metric that rates dichotomies that require proximal task conditions to be placed on opposite sides of the decision boundary as more difficult. Note that proximity of task conditions in task space here is defined with respect to the variables that were manipulated to construct the task space. The conditions corresponding to (response L, outcome low, context 1) and (response L, outcome low, context 2) are proximal because their task specifications differ by a single variable (hamming distance 1) whereas (response L, outcome low, context 1) and (response R, outcome high, context 2) are distal as their task specifications differ by all three variables (Hamming distance 3). With this perspective, we can systematically grade the degree of nonlinearity required to decode a given dichotomy with high accuracy as a function of the number of adjacent task conditions that are on opposite sides of the classification boundary for that dichotomy. For a set of eight conditions specified by three binary variables, this corresponds to the number of adjacent vertices on the cube defined by the variables that are in opposing classes (Extended Data Fig. 5a). We define this number as the difficulty for a given dichotomy, and can compute it directly for every one of the 35 balanced dichotomies. The smallest realizable dichotomy difficulty is 4, and corresponds only to named dichotomies that align with the axis of one of the three binary variables used to specify the task space. The largest realizable dichotomy is 12, and this corresponds to the parity dichotomy because the dichotomy difficulty (number of adjacent conditions with opposing class membership) is maximized in this dichotomy by definition. All remaining dichotomies lie between these two extremes in difficulty, and computing average decoding accuracy over dichotomies of increasing difficulty gives a sensitive readout of the degree of nonlinear task variable interaction present in a neural population.
Geometric analysis of balanced dichotomies
We used three measures to quantify the geometric structure of the neural representation11: shattering dimensionality, CCGP and PS. A high CCGP and PS for a variable indicates that the variable is represented in an abstract format.
Shattering dimensionality is defined as the average decoding accuracy across all balanced dichotomies. It is an index of the expressiveness of a representation, as representations with higher shattering dimensionality allow more dichotomies to be decoded. The content of a representation is assessed by considering which balanced dichotomies are individually decodable better than expected by chance.
CCGP assesses the extent to which training a decoder on one set of conditions generalized to decoding a separate set of conditions. High CCGP for a given variable indicates that the representation of that variable is disentangled from other variables. CCGP is reported in a cross-validated manner by training and testing decoders on single trials. Note that to compute CCGP, all trials from a set of conditions are held out from the training data, which is different from the typical âleave-one-outâ type decoding. The remaining held-in conditions are used to train the decoder, and performance is then evaluated on the held-out conditions (trial-by-trial performance). The CCGP for a given balanced dichotomy is the average over all possible 16 combinations of held-out conditions on either side of the dichotomy boundary. One of the four conditions on each side of the dichotomy are used for testing, whereas the remaining three on each side of the dichotomy are used for training. For each of the 16 possible train and test splits, the decoder is trained on all correct trials from the remaining six conditions, and performance is evaluated on the two held-out conditions.
PS assesses how coding directions for one variable are related to each other across values of other variables in a decoder agnostic manner. The PS is defined for every balanced dichotomy as the cosine of the angle between two coding vectors pointing from conditions in one class to conditions in the other for a given dichotomy. The coding directions are estimated using the average activity for each condition. Note that the PS is a direct geometrical measure that focuses on the structure of the representation, whereas the CCGP also depends on the noise and its shape because it is based on single trials. Coding vectors are computed by selecting four conditions (two on either side of the dichotomy), computing the normalized vector difference between the mean population response for each of the two pairs, then computing the cosine between said coding vectors. This procedure is repeated for all possible pairs of coding vectors, and the average over all cosines is reported. As the correct way of pairing conditions on either side of the dichotomy is not known a priori, we compute the cosine average for all possible configurations of pairing conditions on either side of the dichotomy, then report the PS as the maximum average cosine value over configurations.
Null distribution for geometric measures
We used two approaches to construct null distributions for significance testing of the geometric measures shattering dimensionality, CCGP and PS.
For the shattering dimensionality and decoding accuracy of individual dichotomies, the null distribution was constructed by shuffling trial labels between the two classes on either side of each dichotomy before training and testing the decoder. After shuffling the order of the trial labels, the identical procedures for training and testing were used. This way of constructing the null distribution destroys the information content of the neural population while preserving single-neuron properties such as mean firing rate and variance.
For the CCGP and PA, we used a geometric null distribution11. Before training, we randomly swapped the responses of pairs of neurons within a given condition. For example, for one task condition, all of neuron 1âs responses are assigned to neuron 2 and all of neuron 2âs responses are assigned to neuron 1; for another task condition, all of neuron 1âs responses are assigned to neuron 3, and so on). This way of randomly shuffling entire condition responses leads to the situation in which neural population responses by condition are held constant, but the systematic cross-condition relationships that exist for a given neuron are destroyed. This way of shuffling creates a maximally high-dimensional representation, thereby establishing a conservative null distribution for the geometric measures CCGP and PS.
All null distributions are constructed from 1,000 iterations of shuffled trial-resampling using either trial-label shuffling (shuffle null) or random rotations designed to destroy low-dimensional structure (geometric null).
Neural geometry alignment analysis
To answer the question of whether the geometry of a variable was common across different groups of sessions, we aligned representations between two neural state spaces. Each state space is formed by non-overlapping sets of neurons, and the two spaces are aligned using subsets of task conditions. A cross-session-group PS was then computed by applying the same alignment to a pair of held-out conditions, one on either side of the current dichotomy boundary. Alignment and cross-group comparisons were performed in a space derived using dimensionality reduction (six dimensions). For a given dichotomy, two groups of sessions with N and M neurons were aligned by applying singular value decomposition to the firing-rate normalized condition averages of all but two of the eight task conditions, one on either side of the dichotomy boundary. The top six singular vectors corresponding to the non-zero singular values from each session group were then used as projection matrices to embed the condition averages from each session group in a six-dimensional space. Alignment between the two groups of sessions, in the six-dimensional space, was then performed by computing the average coding vector crossing the dichotomy boundary for each session group, with the vector difference between these two coding vectors defining the âtransformationâ between the two embedding spaces. To compare whether coding directions generalize between the two groups of sessions, we then used the data from the two remaining held-out conditions (in both session groups). We first projected these data points into the same six-dimensional embedding spaces and computed the coding vectors between the two in each embedding space. We then applied the transformation vector to the coding vector in the first embedding space, thereby transforming it into the coordinate system of the second session groups. Within the second session group embedding space, we then computed the cosine similarity between the transformed coding vector from the first session group and the coding vector from the second session group to examine whether the two were parallel (if so, the coding vectors generalize). We repeated this procedure for each of the other three pairs of conditions being the held-out pair, thereby estimating the vector transformation of each pair of conditions independently. The average cosine similarity was then computed over the held-out pairs. All possible configurations of conditions aligned on either side of the dichotomy boundary are considered (24 in this case), and the maximum cosine similarity over configurations is returned as the PS for that dichotomy (plotted as âcross-halfâ in Extended Data Fig. 3z). As a control, we also computed the PS for held-out conditions within the same embedding space without performing cross-session alignment (plotted as âhalf-splitâ in Extended Data Fig. 3z). Note that the differences in both the average PS and the null distribution when comparing within-session and across-session parallelism are expected behaviours and arise from the increased expressive power of the cross-session approach due to fitting transformation vectors in a relatively low-dimensional (six) space. This step is not performed for the within-session control because there is no need to align neural activity to its own embedding space.
MDS
Low-dimensional visualization of neural state spaces was achieved using multi-dimensional scaling (MDS) performed on matrices of condition-averaged neural responses. Pair-wise distances between condition averages were initially computed in N-dimensional neural state space, where N is the number of neurons used to construct the space. Pair-wise distances were then used to compute either a two- or three-dimensional representation of the condition averages using the âmdscaleâ method in MATLAB. In figures in which two different MDS plots are shown side-by-side, canonical correlation analysis was used to align the axes of the two-dimensionally reduced neural state spaces. This approach was necessary because, in general, neural state spaces constructed with different sets of neurons were being compared. We note that we use MDS only to summarize and visualizing high-dimensional neural representations. All conclusions drawn are based on geometric measures computed in the original full neural state space.
Analysis of incorrect trials
For determining decoding accuracy for trials in which participants provided an incorrect response (âerror trialsâ), decoders were trained and evaluated out of sample on all correct trials in inference absent and inference present sessions (denoted as inference absent and inference present trials, respectively). The accuracy of the decoder was then evaluated on the left out error trials in the inference present sessions (denoted as âinference present (error)â trials) that were balanced by task condition. Neurons from sessions without at least one incorrect trial for each of the eight conditions were excluded. We did not estimate CCGP separately for correct and incorrect trials. The PS was estimated using only correct trials for inference present and inference absent. For inference present (error), parallelism was computed using one coding vector (difference between two conditions) from correct trials and one coding vector from incorrect trials. All other aspects of the PS calculation remained as described earlier. The very first trial after a context switch was excluded from analysis (it was incorrect but by design, as the participant cannot know when a context switch occurred).
Stimulus identity geometry analysis
We repeated the geometric analysis described above for subsets of trials to examine specifically how the two variables context and stimulus interact with each other. To do so, we considered each possible pair of stimuli (AB, AC, AD, BC, BD, CD) separately. For each stimulus pair, we then examine the ability to decode and the structure of the underlying representation for two variables: stimulus identity (Supplementary Table 3) and context (Supplementary Table 4) (Fig. 3).
For stimulus identity, what is decoded is whether the stimulus identity is the first or second possible identity in each pair (that is, âA versus Bâ for the AB pair). Stimulus CCGP (Fig. 3b,e) is calculated by training a decoder to decide A versus B in context 1 and testing the decoder in context 2 and vice versa (the CCGP is the average between these two decoders). Stimulus PS (Fig. 3c,f) is the angle between the two coding vectors A versus B in context 1 and 2.
For context, decoding accuracy is estimated by training two decoders to decide context 1 versus context 2 for each of the two stimuli in a stimulus pair. The reported decoding accuracy is the average between these two decoders (Extended Data Fig. 7a,b). For example, for the stimulus pair AB, one such decoder each is trained for all A trials and all B trials. Context CCGP (Fig. 3g and Extended Data Fig. 7c) is calculated by training a decoder to differentiate between contexts 1 and 2 based on the trials in the first identity of the pair and tested in the second pair, and vice versa. The reported context CCGP value for a given stimulus pair is the average between the two. Similarly, context PS (Fig. 3h and Extended Data Fig. 7d) is the angle between the two coding vectors context 1 versus context 2 estimated separately for the first and second stimulus in a pair.
Distance and variance analysis
We computed a series of metrics to quantify aspects of the population response that changed between inference absent and inference present sessions. We used (1) the firing rate, (2) distance in neural state space between classes for balanced dichotomies and stimulus dichotomies (dichotomy distance), (3) variance of neural spiking projected along the coding directions for those dichotomies (coding direction variance) and (4) the condition-wise fano factor (Fig. 4).
Firing rate (Fig. 4e) was the mean firing rate averaged across all neurons during the stimulus period, reported separately for correct trials of every unique task condition. Values reported during the baseline (Extended Data Fig. 9q,r) are computed with an identical procedure using firing rates from before 1âs before stimulus onset.
Dichotomy distance (Fig. 4g,h,j,k) was defined as the Euclidean distance in neural state space between the centroids of the two classes on either side of the decision boundary for that dichotomy. Centroids were computed by constructing the average response vector for each class using a balanced number of correct trials from every condition included in each class through a resampling procedure (described below). Null distributions reported for dichotomy distances are geometric null distributions.
Coding direction variance (Fig. 4i) was computed for a given balanced dichotomy by projecting individual held-out trials onto the coding vector of the decoder trained to differentiate between the two groups of the balanced dichotomy being evaluated. The coding direction was estimated by training a linear decoder on all trials except eight (one from each condition either side of the dichotomy). The vector of weights estimated by the decoder (one for each neuron) was normalized to unit magnitude to estimate the coding vector. The projection of the left out trial onto this coding vector was then calculated using the dot product. This process was repeated 1,000 times, generating a distribution of single-trial projections onto the coding vector for each dichotomy. The variance of the distribution of 1,000 projected data points was then computed and reported as the variance for a given balanced dichotomy (Fig. 4i).
The condition-wise fano factor (Fig. 4f) was computed separately for each neuron. We used all correct trials for a given balanced dichotomy to estimate the mean firing rate and standard deviation and then took the ratio between the two to calculate the fano factor for each neuron. Reported fano factors are the average of all fano factors across all neurons from that area and/or behavioural condition. Fano factors are computed by condition because grouping trials across conditions could lead to task variable coding (signal) contaminating the fano-factor measurement, which should ideally only reflect trial-by-trial variation around the mean for roughly Poisson-distributed firing rates.
The context-modulation consistency (Fig. 4l) was also computed separately for each neuron. Context-modulation consistency is the tendency for a neuronâs firing rate to shift consistently (increase or decrease) to encode context across stimuli. For each neuron, it was computed by determining the sign of the difference (±) between the mean firing rate for a given stimulus between the two contexts, and summing the number of stimuli that show the same modulation (either increase or decrease) across the two contexts. This consistency can take on values between 0 (increase in firing rate to encode context for half of the stimuli, decrease in firing rate for the other half) and 4 (either increase or decrease in firing rate for all four stimuli).
Bootstrap resampled estimation of measures and null distributions
All the measures described in the preceding sections were estimated using a trial and neuron-based resampling method. This resampling strategy was used to assure that every measure reported was comparable between a set of conditions by assuring that the same number of neurons and data points were used to train and test classifiers. Metrics were recomputed 1,000 times with resampling and all null distributions were computed with 1,000 iterations of shuffling and recomputing. Plotted boundaries of null distributions correspond to the fifth and 95th percentiles as estimated from the 1,000 repetitions.
A single iteration of the resampling estimation procedure proceeds as follows. For all analyses that involved a comparison of a metric between two behavioural conditions (inference absent versus inference present or session 1 versus session 2), the same number of neurons was included in both conditions by on a region-by-region basis. For a neuron to be included, at least 15 correct trials for each of the eight unique task conditions had to exist (120 correct trials total). Across patients, the number of correct trials per condition varied: minimum 10.9â±â1.3 trials per condition, mean 25.0â±â0.6 trials per condition, maximum 39.6â±â1.2 trials per condition (meanâ±âs.e.m.). After identifying the neurons that met this inclusion criteria, an equal number were randomly sampled from both behavioural conditions. The number of considered neurons was set to the number of neurons available in the smallest group.
When constructing feature matrices for decoding, 15 trials were randomly selected from each unique condition that was included in the given analysis. Trial order was shuffled independently for every neuron within a condition to destroy potential noise correlations between neurons that were simultaneously recorded. For decoding and shattering dimensionality, out-of-sample accuracy was estimated with fivefold cross-validation. For generalization analyses (CCGP), all trials were used in training as performance was evaluated on entirely held-out conditions. For vector-based measures (dichotomy distance, variance, PS), all trials in relevant conditions were used to compute condition centroids. In the case of variance estimation, all trials except one on either side of the dichotomy boundary were used to learn the coding axis, then the held-out trials were projected onto the coding axis. As previously stated, these procedures were repeated 1,000 times with independent random seeds to ensure independent random sampling of neurons and trials across iterations.
Statistics
All significance values (P values) in the paper are estimated as following unless stated otherwise. P values of decodability, CCGP or PS versus chance for the absent and present group are labelled as PAbsent or PPresent, respectively, and are estimated using a one-sided non-parametric bootstrap test based on the empirically estimated null distribution as described above. The P value of the non-parametric boostrap test is equal to the number of iterations in the null distribution that are larger or equal than the observed value (one-sided), divided by the number of iterations. P values for comparing decodability, CCGP or PS between the inference absent and present conditions are performed using a two-sided Wilcoxon rank-sum test and labelled as PRS. Differences between decodability, CCGP or PS between two conditions are tested using the empirically estimated empirically based on the null distribution estimated as described above and labelled as PÎVariable.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.